Other parts: czech.
Uncle John Returns posted the tables below on Twitter and wrote:
"Turns out that those Czechs who received a dose of
Moderna in 2021 were twice as likely to have a pre-existing chronic
condition as those who had Pfizer with AZ higher still. [...] The source is a ministry of health file which
has 1 row per dose (so 3.68 GB) with other
data such as the age band, sex and place of residence. Available on the
vaccinations tab here
https://


The value of the indikace_ column
(indication chronic illness) is always either
1 or empty. I thought that the percentage of people where the value of
the "indication chronic illness" column was 1
was surprisingly low, because there's only one age group where the
column is true for over 10% of people with a first dose, and the
percentage seemed to be fairly high in young age groups relative to old
age groups (and for some reason it's about 10 times
higher in ages 65-69 than ages 80+):
>download. file( " https:// onemocneni- aktualne. mzcr. cz/ api/ v2/ covid- 19/ ockovani- profese. csv", " ockovani- profese. csv") # about 4 GiB >pro=fread( " ockovani- profese. csv") > d=pro[ poradi_ davky==1& vekova_ skupina! =" nezařazeno",.(. N, chronic=sum( indikace_ chronicke_ onemocneni, na. rm=T)),.( type=vakcina, age=as. numeric( sub( "[+-].* ", " ", vekova_ skupina)))] > d[,.( chronicpct=round( sum( chronic)/ sum( N)* 100, 1)), age][ order( age)]| > print( r=F) age chronicpct 0 1. 5 5 0. 0 12 0. 1 16 0. 6 18 1. 3 25 1. 7 30 1. 9 35 2. 4 40 3. 0 45 3. 9 50 5. 1 55 6. 6 60 8. 6 65 11. 1 70 5. 3 75 2. 1 80 1. 1
Clare Craig said that the column didn't mean if the person had a
chronic illness or not but that it referred to "individuals preferentially vaccinated due to chronic
disease", even though she didn't cite any source for why that
would be the case. [https://
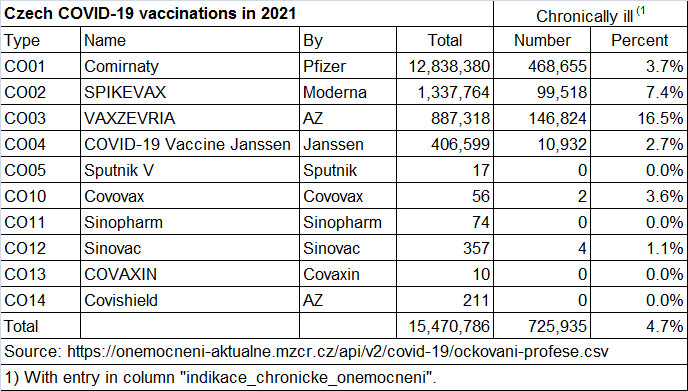
On this page for Czech vaccination statistics, there's 285,232
vaccinated people who have the "indication"
of chronic illness (which is identical to the number
of rows in the ockovani- file where the
indikace_ column is true and the dose
number is 1):
https://
One explanation for why there's so few people who have the indication of chronic illness might be if they only include serious chronic illnesses. But it wouldn't explain why the percentage of people with an indication for chronic illness is much lower in ages 80+ than in ages 65-69. And the list of chronic illnesses in the tooltip also included hypertension, which alone should have a prevalence of more than the total percentage of people with the "indication" of chronic illness.
I found this PDF file which describes the "infectious disease information system" that is
used in the Czech Republic ("Informační systém infekční nemoci", ISIN):
https://
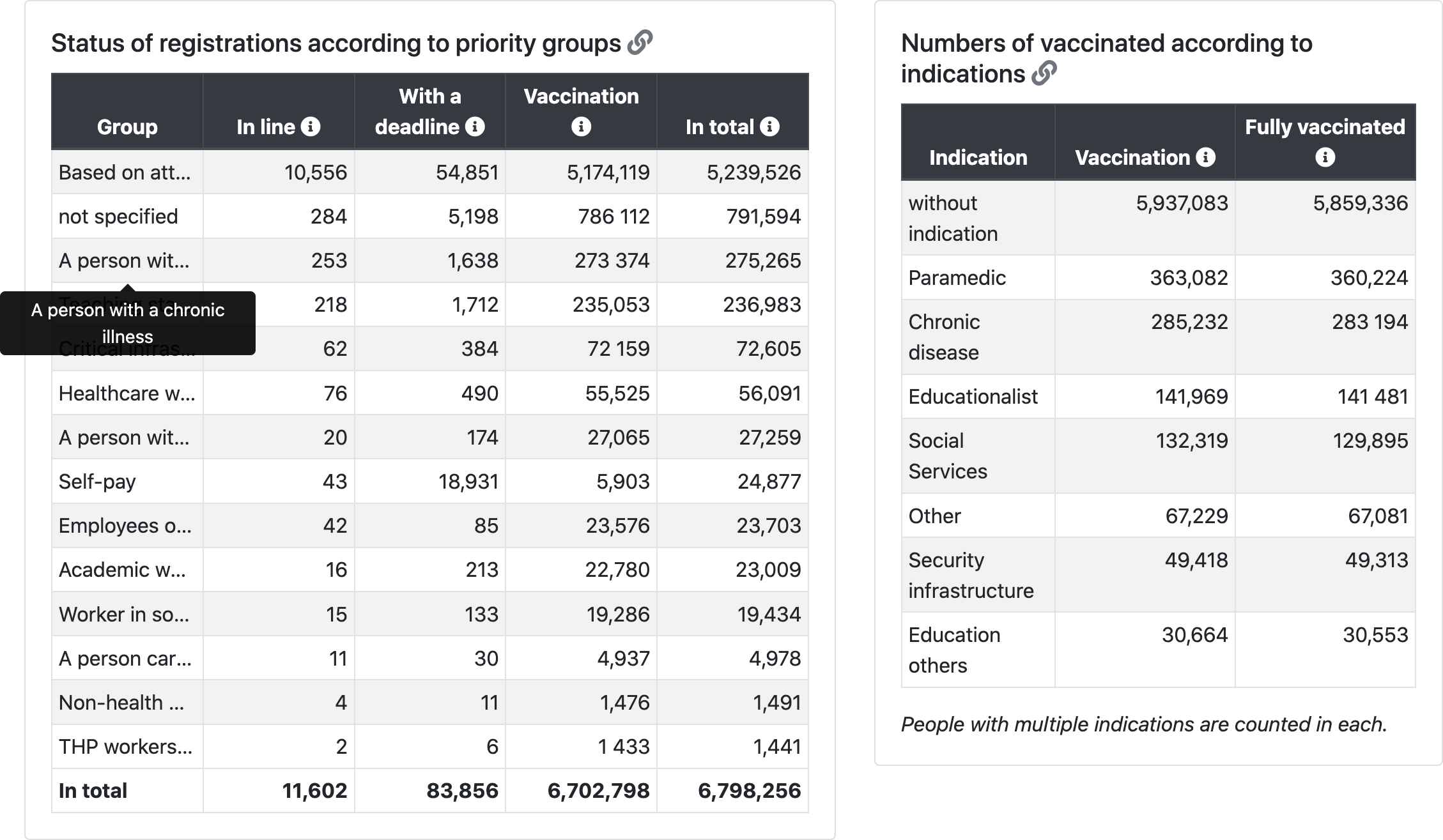
Indications (may change with regard to indications in the Central Reservation System - CRS):
- Professional
- Health workers of the ARO Department, ICU
- Health workers Emergency reception
- Ambulance
- Medical staff of the Infectious Diseases Department
- Medical staff of the Pulmonary Department
- Employees of public health authorities conducting epidemiological investigations
- Laboratory workers processing biological samples for examination on Covid-19
- Workers and clients in social services
- General practitioners, general practitioners for children and adolescents, dentists, pharmacists
- Workers of critical infrastructure - integrated rescue system, energy workers, government, crisis teams
- Other workers of public health protection authorities
- Other healthcare workers
- Employees of the Ministry of Defense
- Teaching staff
- Other workers in education
- Security forces
- Persons involved in the provision of health services
- THP workers in hospitals
- Caregivers (a person caring for a person in the III or IV degree dependencies)
- University academic staff
- Riskiness
- Hemato-oncological disease
- Oncological disease (solid tumors)
- Severe acute or long-term heart disease
- Serious long-term lung disease
- Diabetes mellitus
- Obesity
- Other serious illness
- Severe long-term kidney disease
- Severe long-term liver disease
- Status after transplant or on the waiting list
- Hypertension
- Severe neurological or neuromuscular disease
- Congenital or acquired cognitive deficit
- A rare genetic disease
- Severe weakening of the immune system
- Age
- People aged 80+
- People aged 70-79
- People aged 65-69
- People aged 60-64
- People aged 55-59
- People aged 50-54
- People aged 45-49
- People aged 40-44
- People aged 35-39
- People aged 30-34
- People aged 25-29
- People aged 20-24
- People aged 16-19
- People aged 12-15
- Other
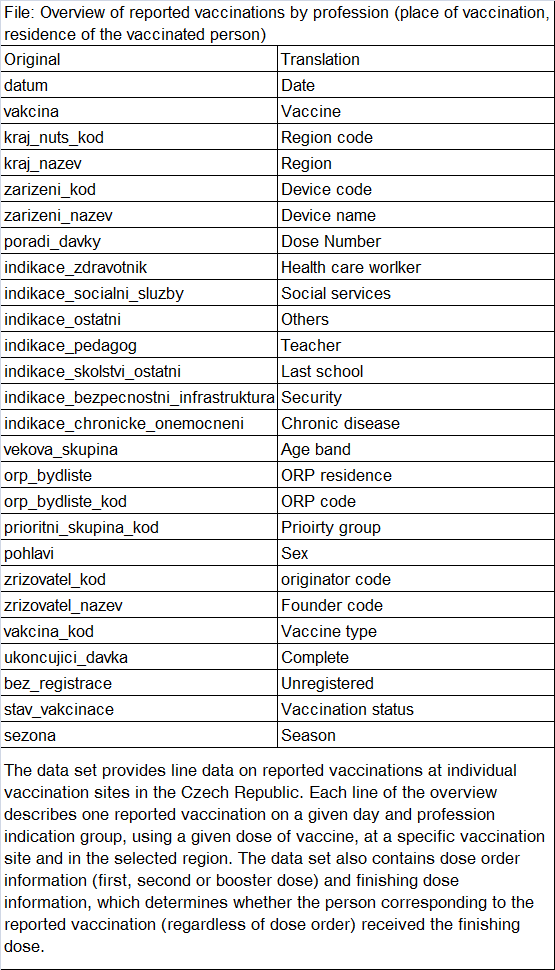
In the file ockovani- which contains one
row for each vaccinated person, there's a column called
povolani (profession) which has
the following values:
| Count | Translation | Czech |
|---|---|---|
| 6037598 | Based on age | Na základě dosaženého věku |
| 912596 | not specified | neuvedeno |
| 305888 | A person with a chronic illness | Osoba s chronickým onemocněním |
| 275259 | Teaching staff/non-teaching staff | Pedagogický pracovník/nepedagogický zaměstnanec |
| 79837 | Critical infrastructure | Kritická infrastruktura |
| 70863 | Healthcare worker according to §76 and §77 of Act 372/2011 Coll. | Zdravotnický pracovník dle §76 a §77 zákona 372/2011 Sb. |
| 39777 | Self-paying | Samoplátce |
| 30231 | A person with a chronic disease - in the care of a specialized center | Osoba s chronickým onemocněním - v péči specializovaného centra |
| 26026 | Employees of the Ministry of Defense | Zaměstnanci Ministerstva obrany |
| 25530 | University academic worker | Akademický pracovník VŠ |
| 21521 | Worker in social services | Pracovník v sociálních službách |
| 5486 | A person caring for a person in 3rd or 4th degree of dependence | Osoba pečující o osobu v III. nebo IV. stupni závislosti |
| 1813 |
Non-health care workers involved in the provision of health care and
care for COVID-19 positive persons |
Nezdravotničtí pracovníci podílející se na poskytování zdravotní
péče a péče o covid-19 pozitivní osoby |
| 1602 | THP workers in hospitals | THP pracovníci v nemocnicích |
The values of the column might indicate the criteria based on which a
person qualified to be vaccinated. By far the most common value of the
column was by age. So if for example a person who was chronically ill
qualified to be vaccinated because of their age, maybe only the
qualification for age but not chronic illness was listed in the column,
since each person has only one value listed in the column. So if a
similar system was used in the ockoni- file, it
might explain why there's such a low percentage of people who are
indicated to have a chronic illness in the oldest age groups.
In the table above the number of people with a chronic illness is
305,888 along with 30,231 people with a chronic illness who were in the
care of a specialized center, so it's a bit higher than the file
ockoni- where there's rows for 285,232 first
doses where the indikace_ column is
true (even though maybe there would be more unique
people where the column would be true for at least one dose, but it's
not possible to tell because the file doesn't have a way to identify
which doses belong to the same person).
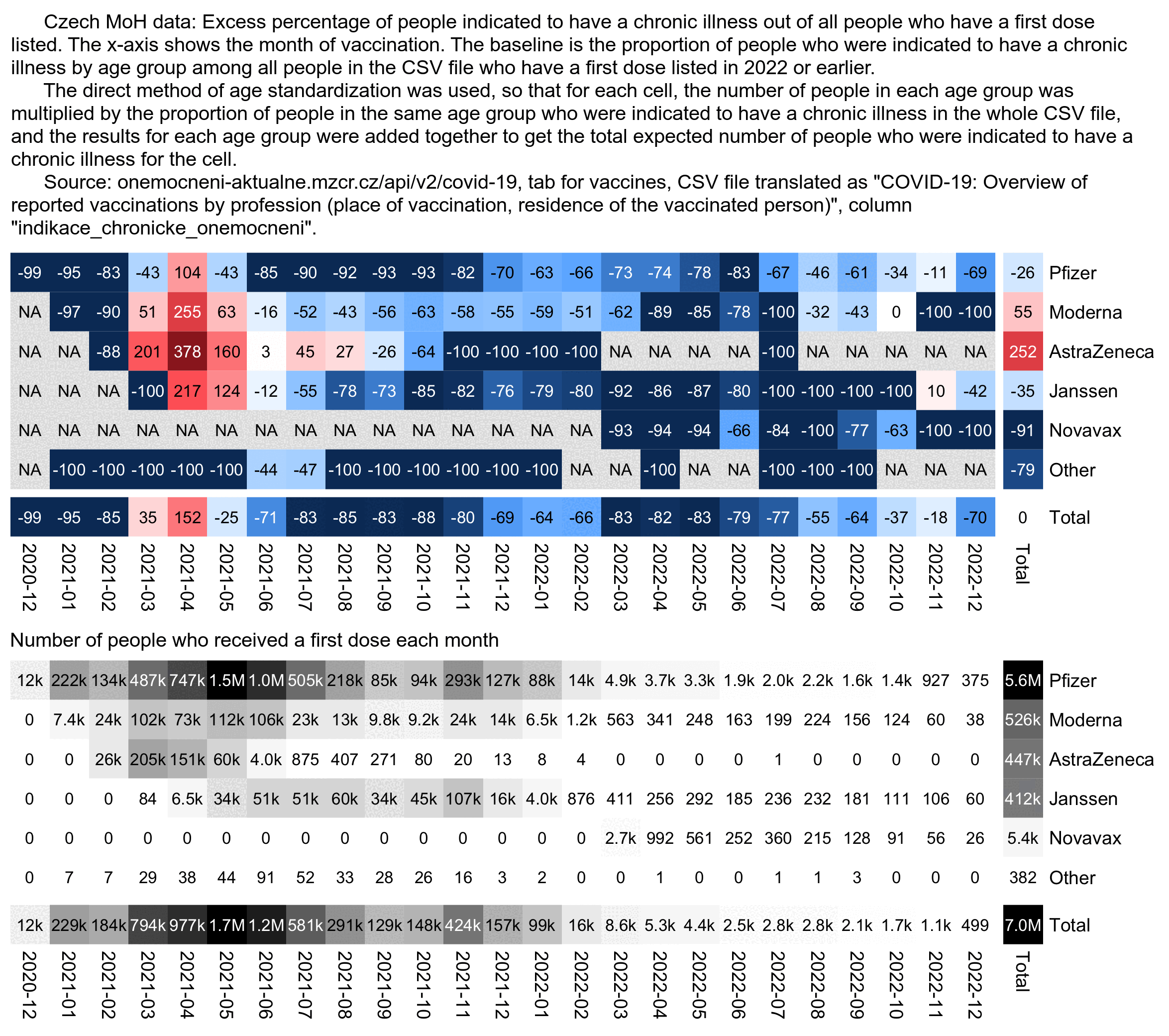
But anyway, in the following code I calculated an age-normalized proportion of people whose "indication chronic illness" column was true, so that my baseline was the proportion of the column by age group among all people who are included in the CSV file. The excess proportion of the column was about -26% for the first dose type Comirnaty but about 55% for SPIKEVAX:
>a=d[,.( N=sum( N), chronic=sum( chronic)),.( age, type)] > a=merge( a, d[,.( base=sum( chronic)/ sum( N)), age])[, base: =base* N] > a[,.( pct=round(( sum( chronic)/ sum( base)- 1)* 100), N=sum( N)), type][ order(- N)]| > print( r=F) type pct N Comirnaty -26 5527776 # Pfizer SPIKEVAX 55 526448 #Moderna VAXZEVRIA 252 447428 #AstraZeneca COVID-19 Vaccine Janssen -35 412320 Comirnaty 5- 11 31 59001 Nuvaxovid -91 5424 # Novavax Comirnaty Omicron XBB.1. 5. 6 1988 Comirnaty Original/ Omicron BA. 4/ BA. 5 -33 669 Comirnaty Original/ Omicron BA. 1 77 294 Sinovac -72 181 Covishield -100 117 Comirnaty 6m- 4 -100 111 Comirnaty Omicron XBB. 1. 5. 5- 11 -31 102 Sinopharm -100 38 COMIRNATY OMICRON XBB. 1. 5 6m- 4 150 29 Covovax -33 29 Spikevax bivalent Original/ Omicron BA. 1 -100 15 Sputnik V -100 10 Nuvaxovid XBB 1. 5 -100 8 COVAXIN -100 5 SPIKEVAX BIVALENT ORIGINAL/ OMICRON BA. 4- 5 310 5 Valneva -100 2 type pct N
The next plot shows the age-normalized proportion of the column by vaccine type and month of vaccination. I was expecting the late vaccinees who got the first dose in late 2021 to have a higher age-normalized proportion of the "indication chronic illness" column than people who got vaccinated during the rollout peak, but actually I got the opposite result. However it might be if the indication for a chronic illness was generally not entered for people who qualified to be vaccinated based on other criteria such as age.
#download. library(file( " https:// onemocneni- aktualne. mzcr. cz/ api/ v2/ covid- 19/ ockovani- profese. csv", " ockovani- profese. csv") data. table) t=fread( " ockovani- profese. csv") yemo=\( x){ u=unique( x); format( u, "% Y-% m")[ match( x, u)]} name1=unique( t$ vakcina); name2=rep( " Other", length( name1)) name2[ name1==" "] =" " name2[ grep( " comirnaty", ignore. case=T, name1)] =" Pfizer" name2[ grep( " spikevax", ignore. case=T, name1)] =" Moderna" name2[ grep( " nuvaxovid", ignore. case=T, name1)] =" Novavax" name2[ name1==" COVID- 19 Vaccine Janssen"] =" Janssen" name2[ name1==" VAXZEVRIA"] =" AstraZeneca" t$ type=name2[ match( t$ vakcina, name1)] d=t[ year( datum) < =2022& poradi_ davky==1,.(. N, chronic=sum( indikace_ chronicke_ onemocneni, na. rm=T)),.( month=yemo( datum), type, age=vekova_ skupina)][ age! =" nezařazeno"] a=d[,.( N=sum( N), chronic=sum( chronic)),.( age, type, month)] a=merge( a, d[,.( base=sum( chronic)/ sum( N)), age])[, base: =base* N] a=a[,.( chronic=sum( chronic), N=sum( N), base=sum( base)),.( type, month)] a[, type: =factor( type, names( sort( a[, tapply( N, type, sum)], T)))] a=rbind( a, a[,.( chronic=sum( chronic), N=sum( N), base=sum( base), month=" Total"), type]) a=rbind( a, a[,.( chronic=sum( chronic), N=sum( N), base=sum( base), type=" Total"), month]) m1=a[, tapply( chronic,.( type, month), c)] m2=a[, tapply( base,.( type, month), c)] pop=a[, xtabs( N~ type+ month)] m=( m1- m2)/ ifelse( m1> m2, m2, m1)* 100 disp=round(( m1/ m2- 1)* 100) exp=. 82; maxcolor=500 m[ m==- Inf] =- maxcolor pheatmap:: pheatmap( abs( m)^ exp* sign( m), filename=" i1. png", display_ numbers=disp, gaps_ row=nrow( m)- 1, gaps_ col=ncol( m)- 1, cluster_ rows=F, cluster_ cols=F, legend=F, cellwidth=19, cellheight=19, fontsize=9, fontsize_ number=8, border_ color=NA, na_ col=" gray90", number_ color=ifelse(( abs( m)^ exp>. 55* maxcolor^ exp) &! is. na( m), " white", " black"), breaks=seq(- maxcolor^ exp, maxcolor^