Other parts: ethical.html, ethical2.html, ethical4.html.
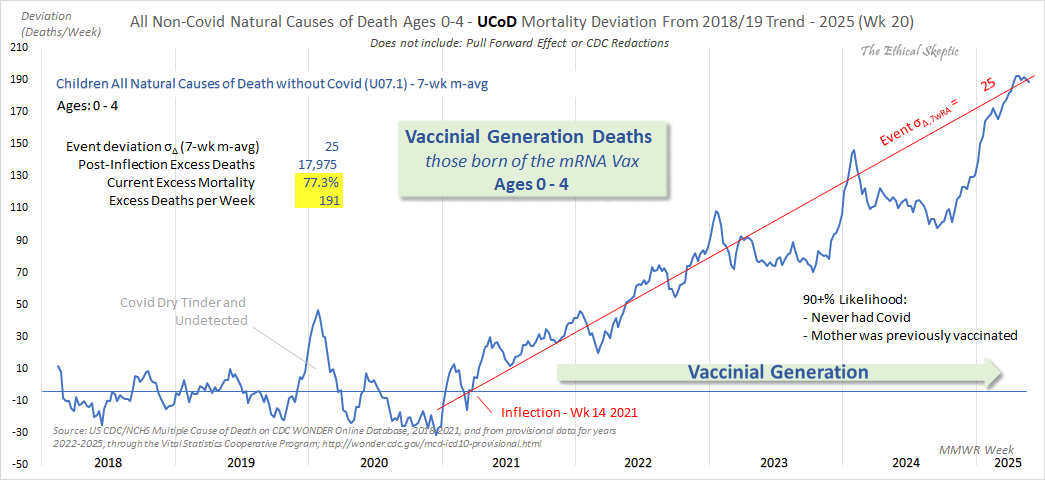
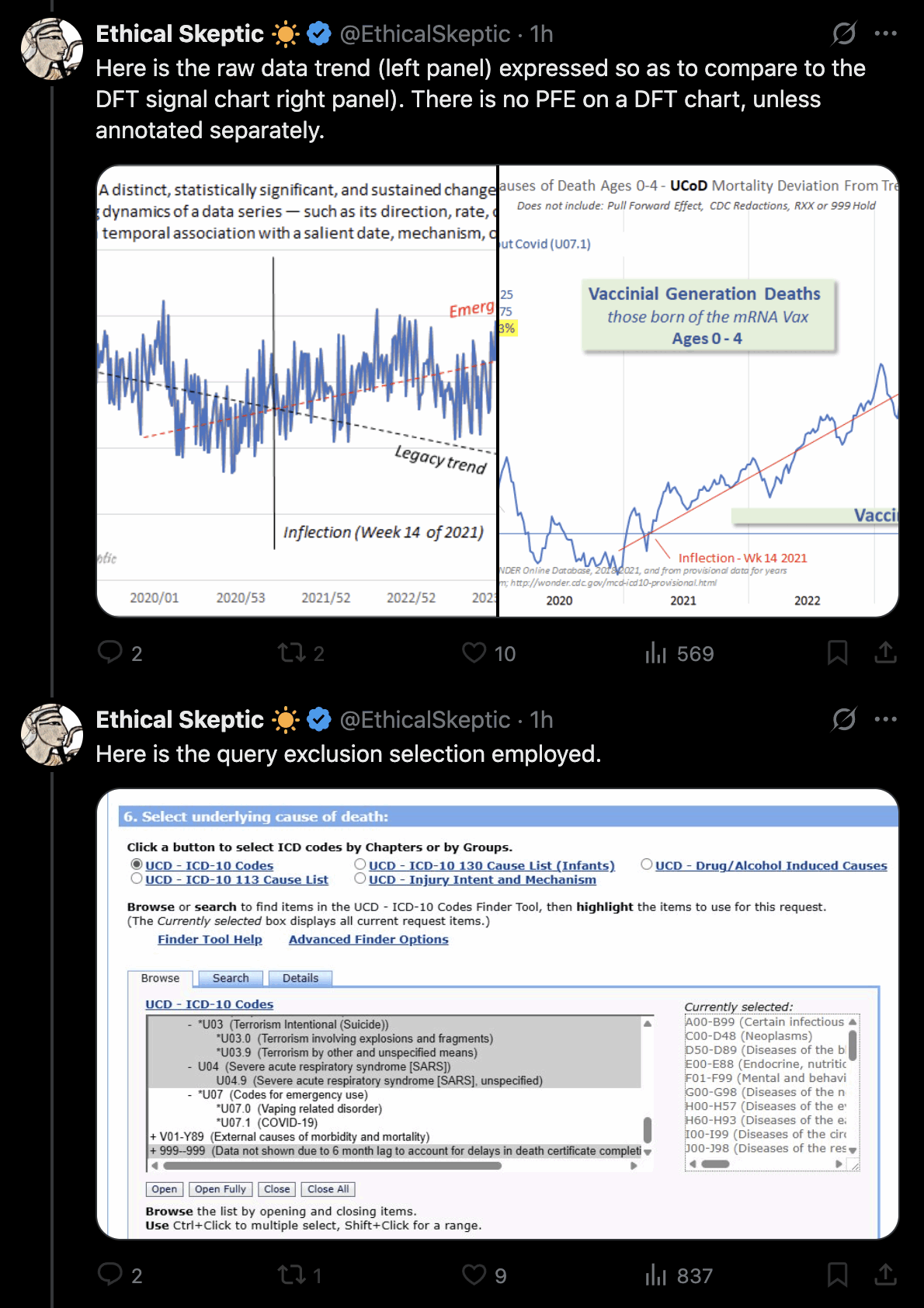
Ethical Skeptic posted this plot of weekly deaths from non-COVID natural causes in ages 0-4, where he got about 77% excess deaths on weeks 14-20 of 2025: [https://web.archive.org/web/20250903223011/https://theethicalskeptic.com/2025/08/19/houston-we-have-another-problem/]

ES has refused to document how he calculated his baseline despite numerous requests. And he hasn't posted any plot that would shown one line for the actual deaths and another line for the baseline.
Therefore I reverse engineered his baseline by digitizing the excess deaths and subtracting them from the actual weekly deaths. ChatGPT and Grok were not successful in digitizing the plot, so I used a tool called WebPlotDigitizer instead:
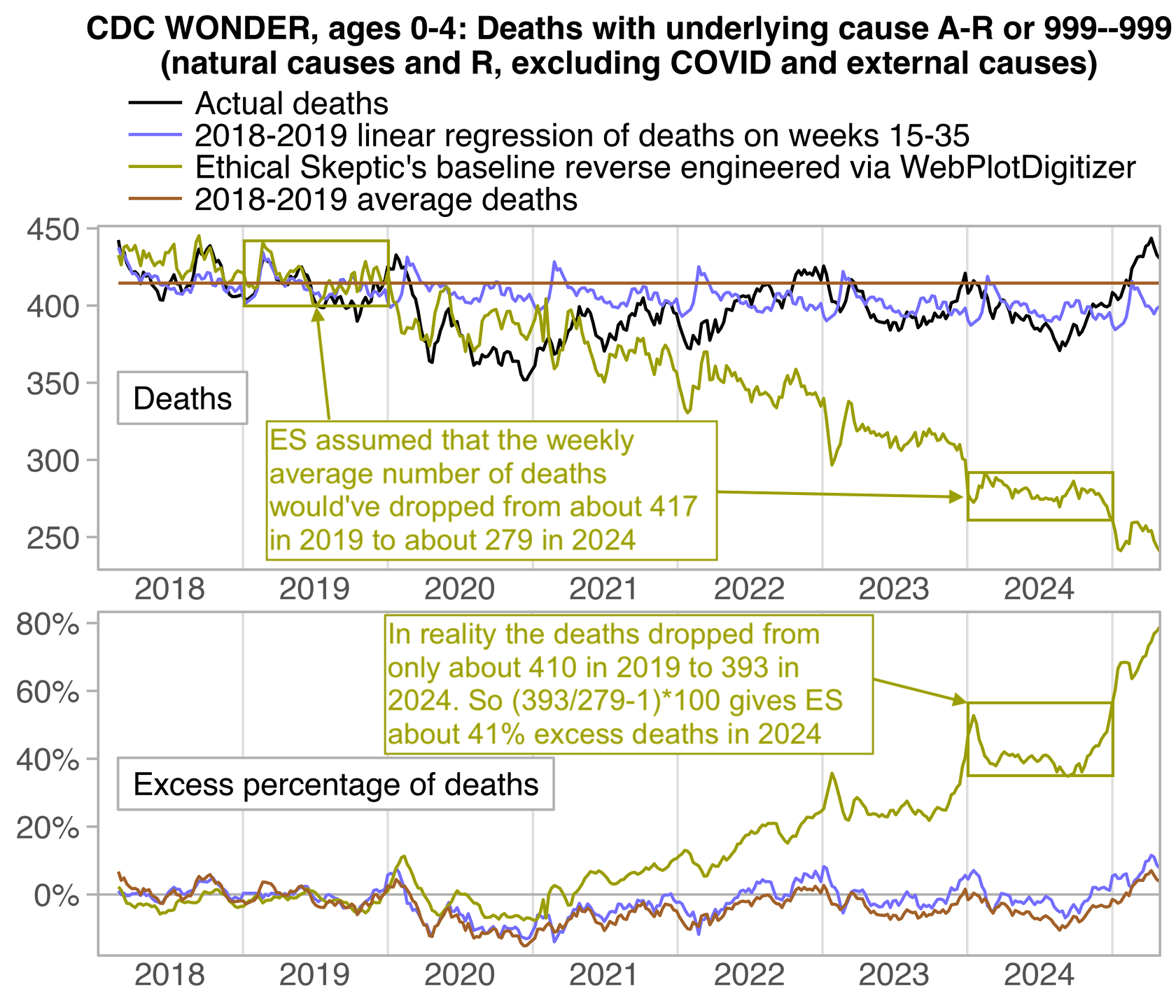
In the next plot the yellow line shows that Ethical Skeptic's baseline roughly followed the slope of the actual deaths between 2018 and 2019, but for some reason his baseline took an unrealistically steep turn downwards between 2019 and 2020. So by 2024 ES gets about 41% excess deaths, because he assumed the deaths would've dropped by about 33% between 2019 and 2024, but in reality the deaths only dropped by about 4% between 2019 and 2024:
ma=\(x,b=1,f=b){x[]=rowMeans(embed(c(rep(NA,b),x,rep(NA,f)),f+b+1),na.rm=T);x}
t=fread("https://sars2.net/f/wondervaccinial0to4.csv")
t[,dead:=ma(dead,3,2)]
t=na.omit(t)
t[,date:=MMWRweek::MMWRweek2Date(year,week,4)]
t=merge(t[year<2020,.(base=mean(dead),base2=mean(dead)),week],t)
slope=t[year<2020,mean(dead),year][,predict(lm(V1~year),.(year=2018:2025))]
slope2=t[year<2020&week%in%15:35,mean(dead),year][,predict(lm(V1~year),.(year=2018:2025))]
t[,base:=base*(slope/mean(slope[1:2]))[factor(t$year)]]
t[,base2:=base2*(slope2/mean(slope2[1:2]))[factor(t$year)]]
t$base3=t[year<2020,predict(lm(dead~date),t)]
t$base3=t$base3+t[year<2020,mean(dead-base3),week]$V1[t$week]
t$ave=t[year<2020,mean(dead)]
lab=c("Actual deaths","2018-2019 linear regression of deaths on weeks 15-35","Ethical Skeptic's baseline reverse engineered via WebPlotDigitizer","2018-2019 average deaths")
p=t[,.(x=date,y=c(dead,base2,dead-excess,ave),z=factor(rep(lab,each=.N),lab))]
p$facet="Deaths"
p=rbind(p,merge(p[z!=z[1]],p[z==z[1],.(x,actual=y)])[,.(x,y=(actual/y-1)*100,z,facet="Excess percentage of deaths")])
p[,facet:=factor(facet,unique(facet))]
xstart=as.Date("2018-1-1");xend=as.Date("2025-5-1")
xbreak=seq(xstart,xend,"6 month");xlab=ifelse(month(xbreak)==7,year(xbreak),"")
ylim=p[,{x=extendrange(y,,.03);.(ymin=x[1],ymax=x[2])},facet]
ggplot(p)+
facet_wrap(~facet,dir="v",scales="free")+
geom_vline(xintercept=seq(xstart,xend,"year"),color="gray90",linewidth=.4)+
geom_segment(data=p[.N],x=xstart,xend=xend,y=0,yend=0,linewidth=.4,color="gray75")+
geom_rect(data=ylim,aes(ymin=ymin,ymax=ymax),xmin=xstart,xmax=xend,lineend="square",linejoin="mitre",fill=NA,color="gray72",linewidth=.4)+
geom_line(aes(x,y,color=z),linewidth=.5)+
geom_label(data=ylim,aes(xstart+50,(ymin+ymax)/2,label=facet),hjust=0,label.r=unit(0,"pt"),label.padding=unit(5,"pt"),label.size=.4,color="gray75",size=3.8)+
geom_label(data=ylim,aes(xstart+50,(ymin+ymax)/2,label=facet),hjust=0,label.r=unit(0,"pt"),label.padding=unit(5,"pt"),label.size=0,size=3.8,fill=NA)+
labs(x=NULL,y=NULL,title="CDC WONDER, ages 0-4: Deaths with underlying cause A-R or 999--999\n(natural causes and R, excluding COVID and external causes)")+
scale_x_continuous(limits=c(xstart,xend),breaks=xbreak,labels=xlab)+
scale_y_continuous(breaks=\(x)pretty(x,7),labels=\(x)if(max(x,na.rm=T)<100)paste0(x,"%")else x)+
scale_color_manual(values=c("black","#8888ff","#aaaa00",hsv(3/36,.7,.7)))+
coord_cartesian(clip="off",expand=F)+
theme(axis.text=element_text(size=11,color="gray40"),
axis.ticks=element_line(linewidth=.4,color="gray75"),
axis.ticks.length.x=unit(0,"pt"),
axis.ticks.length.y=unit(4,"pt"),
legend.background=element_blank(),
legend.box.spacing=unit(0,"pt"),
legend.direction="vertical",
legend.key=element_blank(),
legend.key.height=unit(11,"pt"),
legend.key.width=unit(23,"pt"),
legend.margin=margin(,,4),
legend.position="top",
legend.spacing.x=unit(2,"pt"),
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=11,vjust=.5),
legend.title=element_blank(),
panel.background=element_blank(),
panel.border=element_rect(color="gray75",fill=NA,linewidth=.4),
panel.spacing=unit(2,"pt"),
plot.title=element_text(size=11,face=2,hjust=.5,margin=margin(,,2)),
strip.background=element_blank(),
strip.text=element_blank())
ggsave("1.png",width=5.6,height=4.8,dpi=300*4)
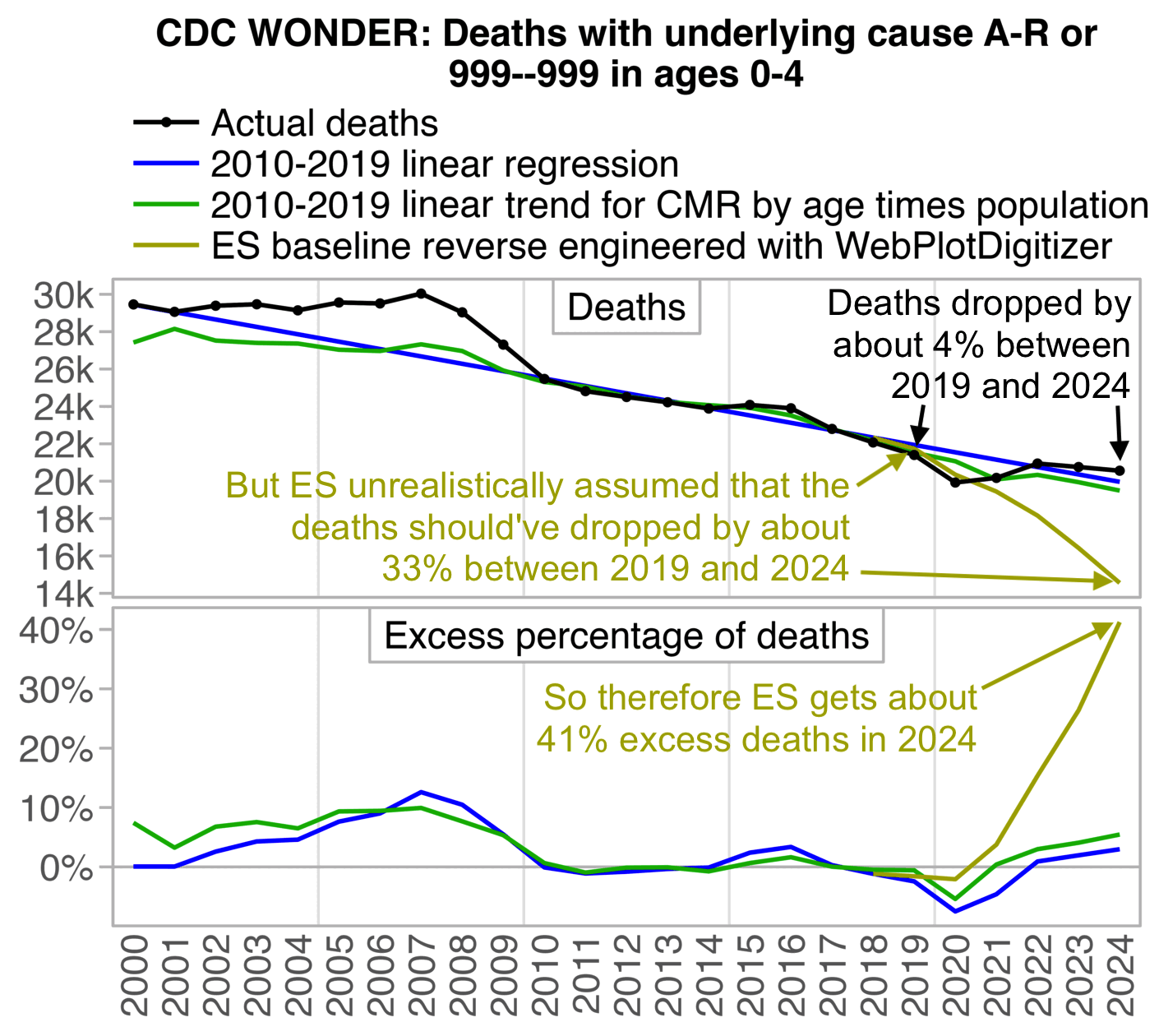
In the next plot I converted Ethical Skeptic's baseline from weekly to yearly deaths. I took the average baseline value on each MMWR year, divided it by 7, and multiplied it by the number of days in the year. Now you can see even more clearly how his baseline matched the pre-COVID trend between 2018 and 2019, but his baseline took an unrealistic dive downwards after 2019:
eth=fread("https://sars2.net/f/wondervaccinial0to4.csv")
eth=na.omit(eth)[,.(eth=mean(dead-excess)/7*(365+(year%%4==0))),year][year<2025]
t=fread("https://sars2.net/f/wondervaccinial0to4ator999monthly.csv")
t=t[age<5&year%in%2000:2024,.(dead=sum(dead)),.(year,age)]
t=merge(t,fread("https://sars2.net/f/uspopdead.csv")[,.(pop,age,year)])
a=merge(t[,.(dead=sum(dead)),year],eth,all=T)
a$base=a[year%in%2010:2019,predict(lm(dead~year),a)]
a$base2=t[,.(year,base=predict(lm(dead/pop~year,.SD[year%in%2010:2019]),.SD)*pop),age][,tapply(base,year,sum)]
p=a[,.(x=year,y=c(dead,base,base2,eth),z=rep(1:4,each=.N),facet=1)]
p=rbind(p,p[,.(x,y=(y[z==1]/y-1)*100,z,facet=2)][z!=1])
p[,z:=factor(z,,c("Actual deaths","2010-2019 linear regression","2010-2019 liner trend for CMR by age times population","ES baseline reverse engineered with WebPlotDigitizer"))]
p[,facet:=factor(facet,,c("Deaths","Excess percentage of deaths"))]
xstart=min(p$x);xend=max(p$x)
ylim=p[,{x=extendrange(y);.(ymin=x[1],ymax=x[2])},facet]
ggplot(p)+
facet_wrap(~facet,dir="v",scales="free_y")+
geom_vline(xintercept=seq(xstart-.5,xend,5),color="gray90",linewidth=.4)+
geom_rect(data=ylim,aes(ymin=ymin,ymax=ymax),xmin=xstart-.5,xmax=xend+.5,lineend="square",linejoin="mitre",fill=NA,color="gray75",linewidth=.4)+
geom_segment(data=ylim[2],y=0,yend=0,x=xstart-.5,xend=xend+.5,linewidth=.4,color="gray75")+
geom_line(aes(x,y,color=z),linewidth=.6)+
geom_line(data=p[z==z[1]],aes(x,y,color=z),linewidth=.6)+
geom_point(aes(x,y,alpha=z,color=z),stroke=0,size=1.4)+
geom_label(data=ylim,aes(label=facet,y=ymax),x=(xstart+xend)/2,hjust=.5,vjust=1,fill="white",label.r=unit(0,"pt"),label.padding=unit(4,"pt"),label.size=.4,color="gray75",size=3.87,lineheight=.8)+
geom_label(data=ylim,aes(label=facet,y=ymax),x=(xstart+xend)/2,hjust=.5,vjust=1,fill=NA,label.r=unit(0,"pt"),label.padding=unit(4,"pt"),label.size=0,size=3.87,lineheight=.8)+
labs(x=NULL,y=NULL,title="CDC WONDER: Deaths with underlying cause A-R or\n999--999 in ages 0-4")+
scale_x_continuous(limits=c(xstart-.5,xend+.5),breaks=xstart:xend)+
scale_y_continuous(breaks=\(x)pretty(x,7),labels=\(x)if(max(x,na.rm=T)<1e3)paste0(x,"%")else paste0(x/1e3,"k"))+
scale_color_manual(values=c("black","blue",hsv(1/3,1,.7),"#aaaa00"))+
scale_alpha_manual(values=c(1,0,0,0,0,0))+
coord_cartesian(clip="off",expand=F)+
theme(axis.text=element_text(size=11,color="gray40"),
axis.text.x=element_text(angle=90,vjust=.5,hjust=1),
axis.text.y=element_text(margin=margin(,1.5)),
axis.ticks=element_line(linewidth=.4,color="gray75"),
axis.ticks.length=unit(4,"pt"),
axis.ticks.length.x=unit(0,"pt"),
legend.background=element_blank(),
legend.box.spacing=unit(0,"pt"),
legend.direction="vertical",
legend.key=element_blank(),
legend.key.height=unit(12,"pt"),
legend.key.width=unit(24,"pt"),
legend.margin=margin(-2,,4),
legend.position="top",
legend.spacing.x=unit(1,"pt"),
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=11,vjust=.5),
legend.title=element_blank(),
panel.background=element_blank(),
panel.border=element_rect(linewidth=.3,fill=NA,color="gray75"),
panel.grid.major=element_blank(),
panel.spacing=unit(3,"pt"),
plot.title=element_text(size=11,face=2,margin=margin(,,4),hjust=.5),
strip.background=element_blank(),
strip.text=element_blank())
ggsave("1.png",width=4.7,height=4.2,dpi=300*4)
Ethical Skeptic's plot says that the current 7-week moving average of excess mortality is 191 deaths per week, or about 77.3% excess deaths. It refers to the excess mortality on MMWR weeks 14 to 20 of 2025:
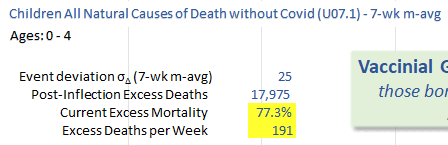
By solving x+191=1.773*x, you get a value of about 247.1
for the baseline number of deaths. It's off by only about 1 death from
the average value of my reverse engineered baseline on weeks 14 to 20 of
2025:
t=fread("https://sars2.net/f/wondervaccinial0to4.csv")
t[year==2025&week%in%14:20,mean(dead-excess)]
# 246.1207
ES posted this screenshot that showed what ICD codes were included in his plot: [https://theethicalskeptic.com/2025/08/19/houston-we-have-another-problem/]
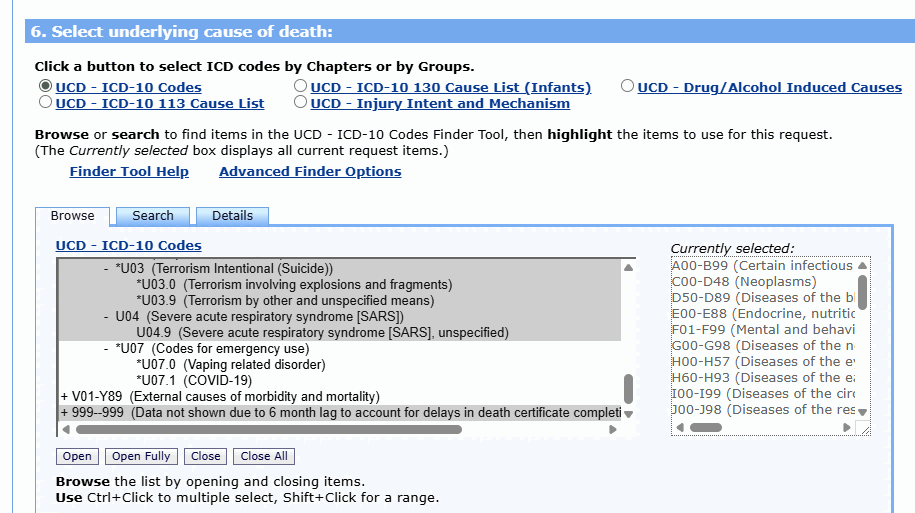
The U chapter consists of ICD codes for terrorism (U01-03), SARS (U04), vaping-related disorder (U07.0), COVID (U07.1), and long COVID (U09). U09 is only used as a multiple cause code and not an underlying cause code. Apart from U07.1 deaths, ages 0-4 had only 4 deaths in 1999-2025 with the underlying cause under the U chapter, so it doesn't make much difference if you exclude the entire U chapter or only U07.1. There's one death in 2022 with the UCD U04.9 (SARS), which might have been a miscoded COVID death. And there's 3 deaths in September 2001 with the UCD U01.1 (terrorism involving destruction of aircraft).
ES seems to have also included R codes in his plot, even though you can't see from his screenshot if he included R codes or not. Out of deaths in ages 0-4 in 2018-2025 with an R code as an underlying cause, about 97% of all deaths had the underlying cause R99 ("Other ill-defined and unspecified causes of mortality") or R95 ("Sudden infant death syndrome - SIDS"):
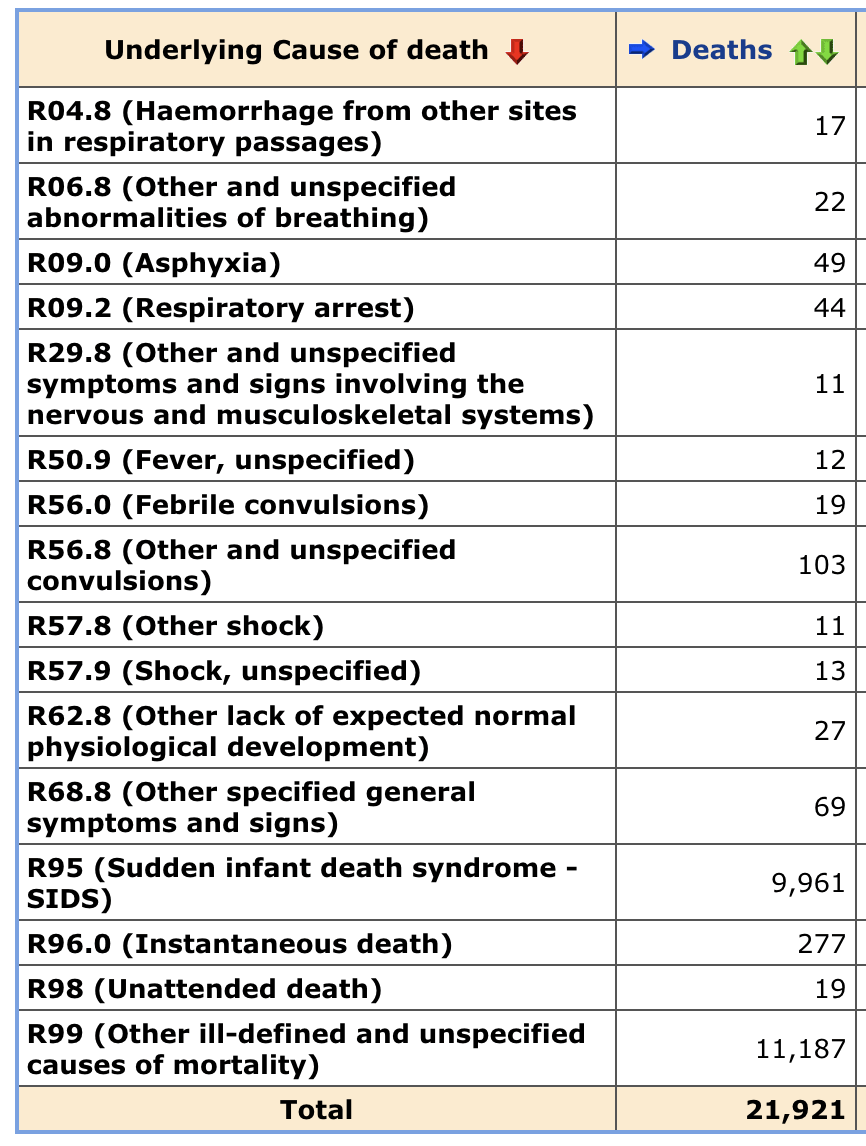
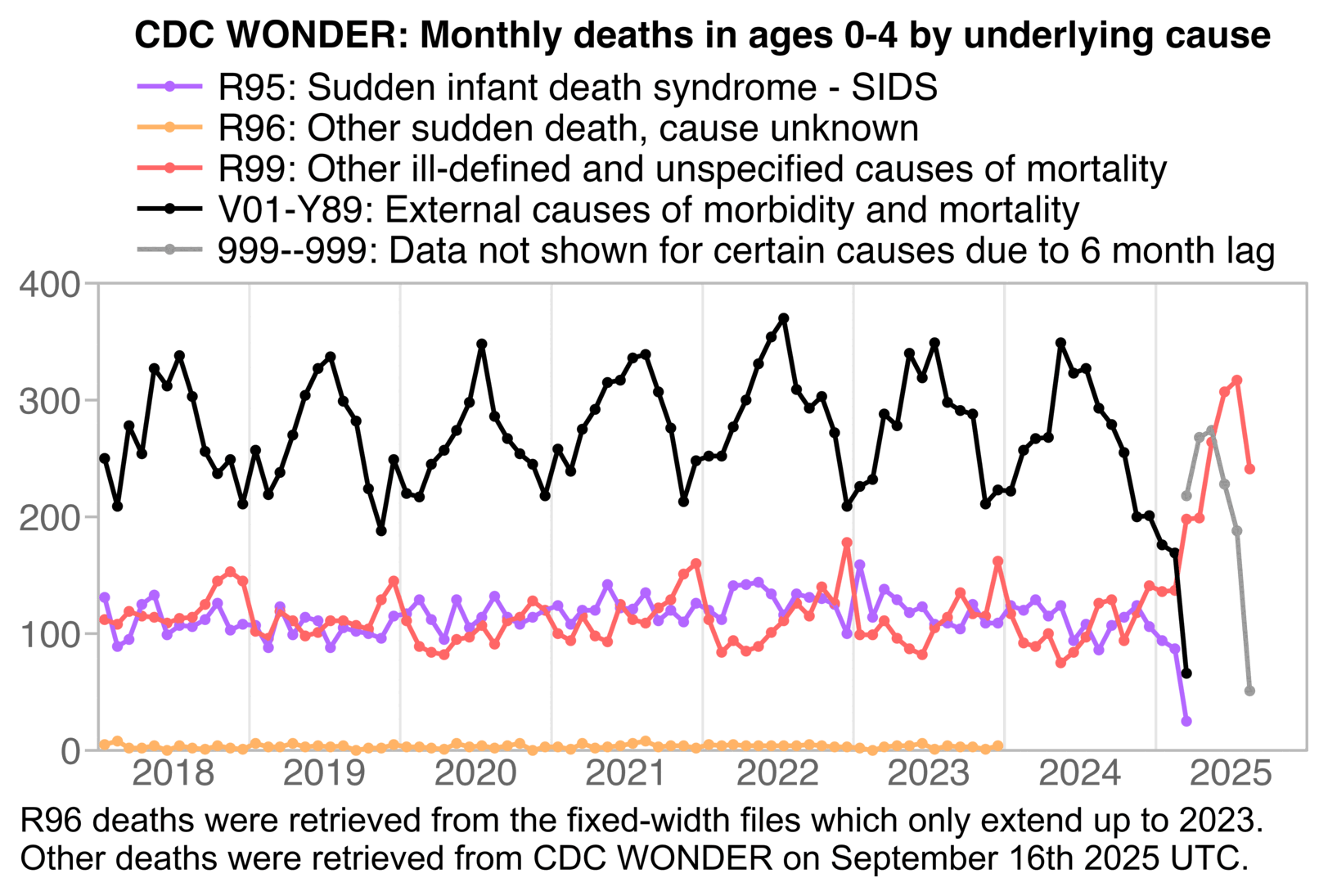
R99 and R96 deaths are elevated in recent months, because R99 and R96 are used as temporary placeholder codes for deaths that will later be assigned under a specific cause. However R95 (SIDS) doesn't appear to be used as a similar placeholder code, and SIDS deaths are not elevated in recent months like R99 deaths.
The documentation at CDC WONDER says that deaths from external causes and SIDS are listed under 999--999 for 24 weeks prior to the final date in the provisional data, which in turn is about 4 weeks before the current week: "In the provisional mortality data, causes of death classified as external causes of injury (ICD-10 codes V01-Y89), sudden deaths (ICD-10 codes R95 and R96), or drug poisoning (ICD-10 codes T36-T50), have the cause of death labeled as 'Data not shown due to 6 month lag to account for delays in death certificate completion for certain causes of death.' For these deaths occurring 24 weeks prior to the final date in the provisional data, the number of deaths and related statistics are reported as Not Available." [https://wonder.cdc.gov/wonder/help/mcd-provisional.html]
When I did a query at CDC WONDER on MMWR week 38 of 2025, the final date included in the provisional data was on week 34, so deaths from external causes and SIDS were only returned up to week 10, and 999--999 deaths were returned from week 11 up to week 34.
Deaths from external causes are also listed under 999--999 for the last 24 weeks of the provisional data, so in fact Ethical Skeptic's plot includes deaths from external causes for several weeks at the end of the x-axis. An earlier version of his plot extended up to week 15 of 2020, but on this page I have written about a version of his plot that extends up to week 20 of 2025, which he first posted around MMWR week 34 of 2025. [https://x.com/EthicalSkeptic/status/1957560098366042437/photo/1] But if on week 34 the last week included in the provisional data was week 30, then deaths from external causes would've been listed under 999--999 from week 7 onwards.
The next plot also demonstrates that when I did a query at CDC WONDER in September 2025, deaths from external causes and SIDS were only returned up to March 2025:
t=fread("https://sars2.net/f/wonder0to4ucdr.csv")
t[code=="999--999",cause:="Data not shown for certain causes due to 6 month lag"]
v=fread("xz -dc https://sars2.net/f/vital.csv.xz")
t2=v[age<5][year>2017&cause%like%"R96",.(dead=sum(ucd)),.(year,month)]
t2=merge(t2,do.call(CJ,lapply(t2[,1:2],unique)),all=T)[is.na(dead),dead:=0]
t2[,code:="R96"][,cause:="Other sudden death, cause unknown"]
t=rbind(t,t2)
t=rbind(t[order(code)][code!="999--999"],t[code=="999--999"])
t[,z:=paste0(code,": ",cause)][,z:=factor(z,unique(z))]
p=t[,.(x=as.Date(paste(year,month,16,sep="-")),y=dead,z)]
xstart=as.Date("2018-1-1");xend=as.Date("2026-1-1");xbreak=seq(xstart+182,xend,"year")
ybreak=pretty(c(0,p$y));ystart=0;yend=max(ybreak)
ggplot(p)+
geom_vline(xintercept=seq(xstart,xend,"year"),color="gray90",linewidth=.4)+
annotate("rect",xmin=xstart,xmax=xend,ymin=ystart,ymax=yend,linewidth=.4,lineend="square",linejoin="mitre",fill=NA,color="gray75")+
geom_point(aes(x,y,color=z),stroke=0,size=1.5)+
geom_line(aes(x,y,color=z),linewidth=.65)+
labs(x=NULL,y=NULL,title="CDC WONDER: Monthly deaths in ages 0-4 by underlying cause")+
scale_x_continuous(limits=c(xstart,xend),breaks=xbreak,labels=year(xbreak))+
scale_y_continuous(limits=c(ystart,yend),breaks=ybreak)+
scale_color_manual(values=c(hsv(3/4,.6,1),hsv(1:0/12,.6,1),"black","gray60"))+
coord_cartesian(clip="off",expand=F)+
guides(color=guide_legend(ncol=1,byrow=F))+
theme(axis.text=element_text(size=11,color="gray40"),
axis.text.y=element_text(margin=margin(,1)),
axis.ticks=element_line(linewidth=.4,color="gray75"),
axis.ticks.length=unit(0,"pt"),
axis.ticks.length.y=unit(4,"pt"),
legend.background=element_blank(),
legend.box.spacing=unit(0,"pt"),
legend.direction="vertical",
legend.key=element_blank(),
legend.key.height=unit(12,"pt"),
legend.key.width=unit(24,"pt"),
legend.margin=margin(-1,,4),
legend.position="top",
legend.spacing.x=unit(2,"pt"),
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=11),
legend.title=element_blank(),
panel.background=element_blank(),
plot.title=element_text(size=11,hjust=.5,face=2,margin=margin(,,4)))
ggsave("1.png",width=5.4,height=3.3,dpi=300*4)
So R99 and 999--999 inflate the deaths in Ethical Skeptic's plot in 2025, because many of the deaths that are temporarily assigned under R99 or 999--999 will later be assigned under external causes. If he would've simply plotted deaths from all causes, then he could've avoided the bias caused by R99 and 999--999 deaths.
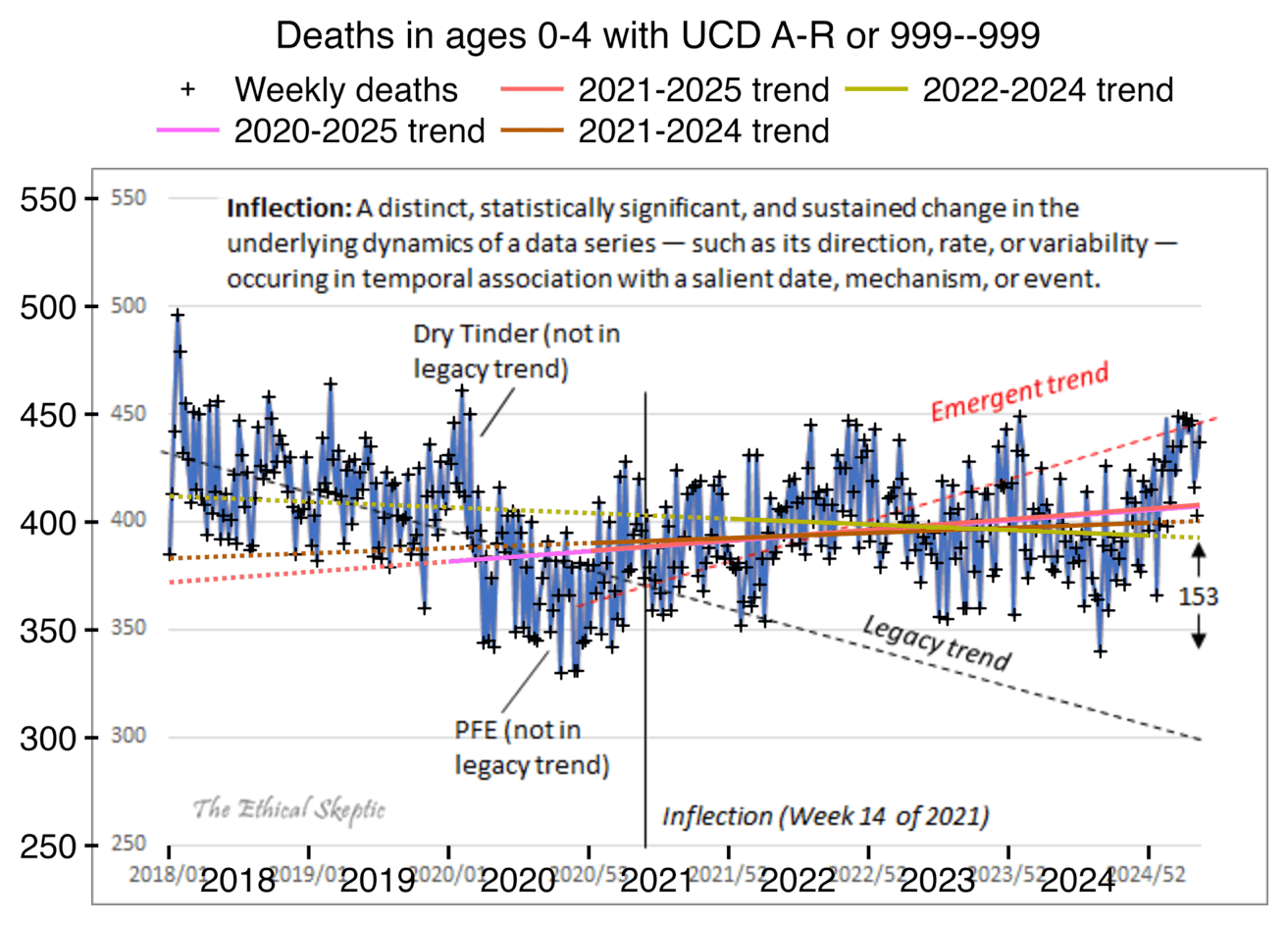
ES included this plot in his blog post: [https://theethicalskeptic.com/2025/08/19/houston-we-have-another-problem/]
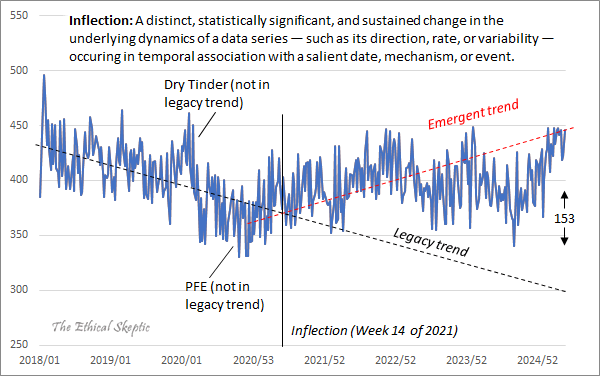
The "legacy trend" looks far too steep, because it appears to approximate the trend in 2018-2020, but there was an unusually low number of deaths in 2020. If 2020 or later years are included in the baseline fitting period, ES says that it is a grave sin that he has termed "paltering". But he now seems to have succumbed to the sin of paltering himself.
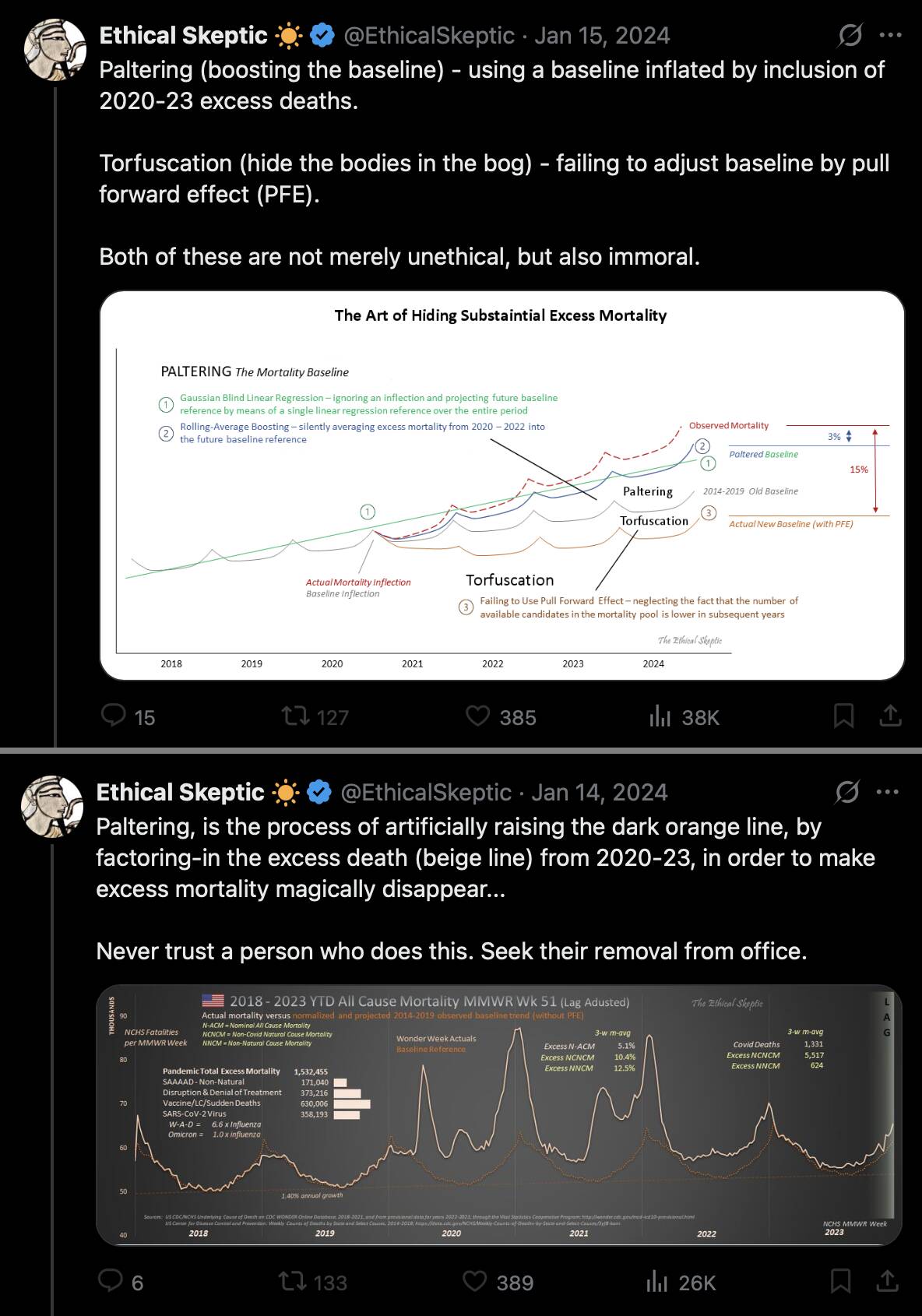
ES said that paltering is not merely unethical but also immoral, and that a person who commits paltering should be removed from office: [https://x.com/EthicalSkeptic/status/1746685316465852776, https://x.com/EthicalSkeptic/status/1746594438174978431]
In the next plot I fitted the two pink baselines against weekly deaths in 2018-2019, and I fitted the two green baselines against monthly deaths in 2013-2019. But none of my four baselines are anywhere near as steep as Ethical Skeptic's legacy trend:
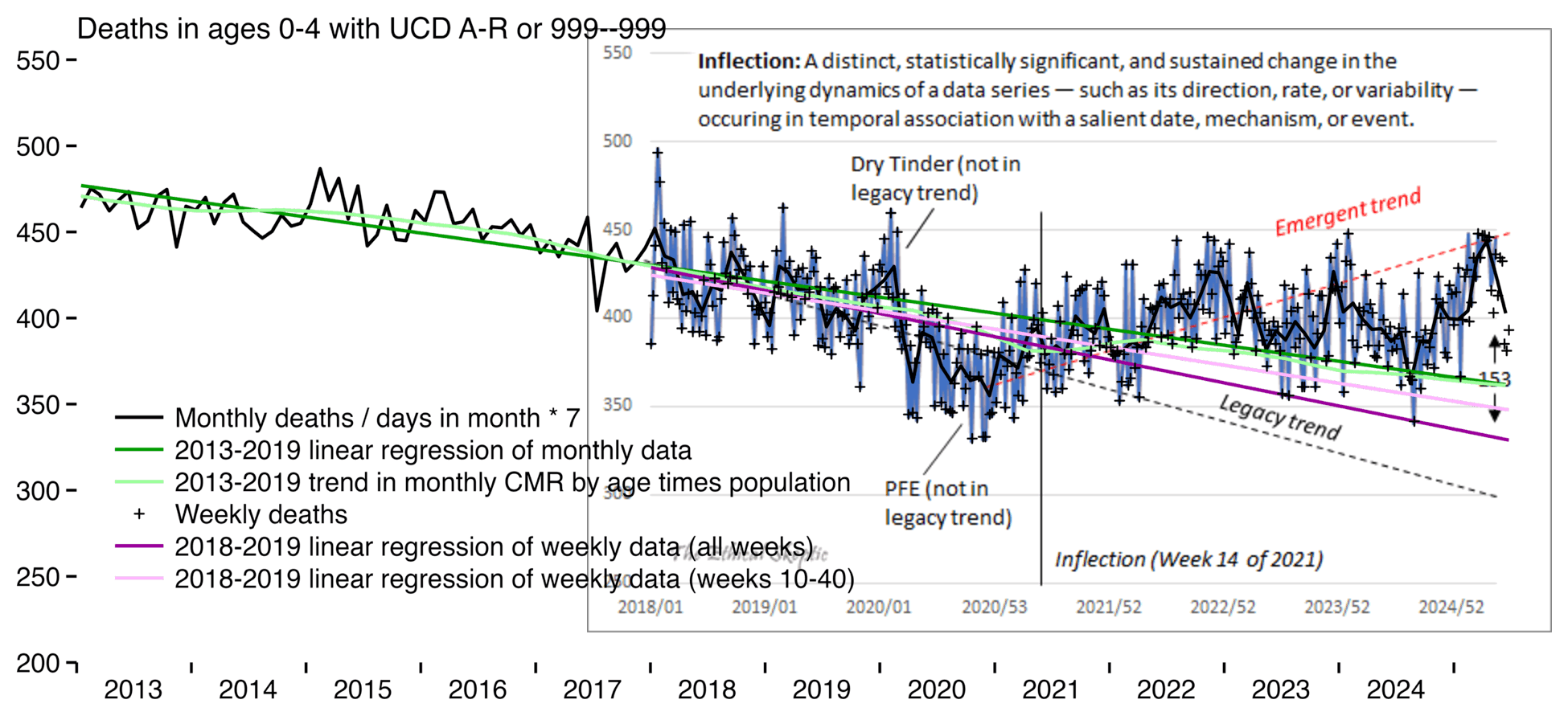
t=fread("https://sars2.net/f/wondervaccinial0to4ator999monthly.csv")[age<5]
t=merge(t,fread("https://sars2.net/f/uspopdeadmonthly.csv")[,.(year,month,age,pop)])[year>=2013]
t[,x:=as.Date(paste(year,month,15,sep="-"))]
t[,dead:=dead/days_in_month(x)*7]
t=merge(t[,.(x,base=predict(lm(dead/pop~x,.SD[year<2020]),.SD)),age],t)
d=t[,.(dead=sum(dead),base=sum(base*pop)),x]
d$base2=d[year(x)<2020,predict(lm(dead~x),d)]
p=d[,.(x,y=c(dead,base2,base),z=rep(1:3,each=.N))]
t=fread("https://sars2.net/f/wondervaccinial0to4.csv")
t[,x:=MMWRweek::MMWRweek2Date(year,week,4)]
t$base=t[year<2020,predict(lm(dead~x),t)]
t$base2=t[year<2020&week%in%10:40,predict(lm(dead~x),t)]
p=rbind(p,t[,.(x,y=c(dead,base,base2),z=rep(4:6,each=.N))])
p[,z:=factor(z,,c("Monthly deaths / days in month * 7","2013-2019 linear regression of monthly data","2013-2019 trend in monthly CMR by age times population","Weekly deaths","2018-2019 linear regression of weekly data (all weeks)","2018-2019 linear regression of weekly data (weeks 10-40)"))]
xstart=as.Date("2013-1-1");xend=as.Date("2025-7-1");xbreak=seq(xstart,xend-1,"6 month")
xlab=ifelse(month(xbreak)==7,year(xbreak),"")
ystart=200;yend=550;ybreak=0:11*50
ggplot(p)+
geom_line(aes(x,y,color=z,linewidth=z))+
geom_point(aes(x,y,color=z,alpha=z),shape=3,stroke=.5,size=.8)+
labs(title="Deaths in ages 0-4 with UCD A-R or 999--999",x=NULL,y=NULL)+
scale_x_continuous(limits=c(xstart,xend),breaks=xbreak,labels=xlab)+
scale_y_continuous(limits=c(ystart,yend),breaks=ybreak)+
scale_color_manual(values=c("black",hsv(12/36,c(1,.4),c(.6,1)),"black",hsv(30/36,c(1,.3),c(.6,1))))+
scale_linewidth_manual(values=c(.6,.6,.6,0,.6,.6))+
scale_alpha_manual(values=c(0,0,0,1,0,0))+
coord_cartesian(clip="off",expand=F)+
theme(axis.text=element_text(size=10,color="black"),
axis.ticks=element_line(color=alpha("black")),
axis.ticks.length=unit(4,"pt"),
axis.ticks.x=element_line(color=alpha("black",c(1,0))),
legend.background=element_blank(),
legend.box.spacing=unit(0,"pt"),
legend.justification=c(0,0),
legend.key=element_blank(),
legend.key.height=unit(12,"pt"),
legend.key.width=unit(23,"pt"),
legend.margin=margin(3,5,3,3),
legend.position=c(.017,.1),
legend.spacing.x=unit(2,"pt"),
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=10,vjust=.5),
legend.title=element_blank(),
panel.background=element_blank(),
panel.grid=element_blank(),
plot.background=element_rect(fill="transparent",color=NA),
plot.title=element_text(size=11))
ggsave("1.png",width=8,height=2.7,dpi=300*4)
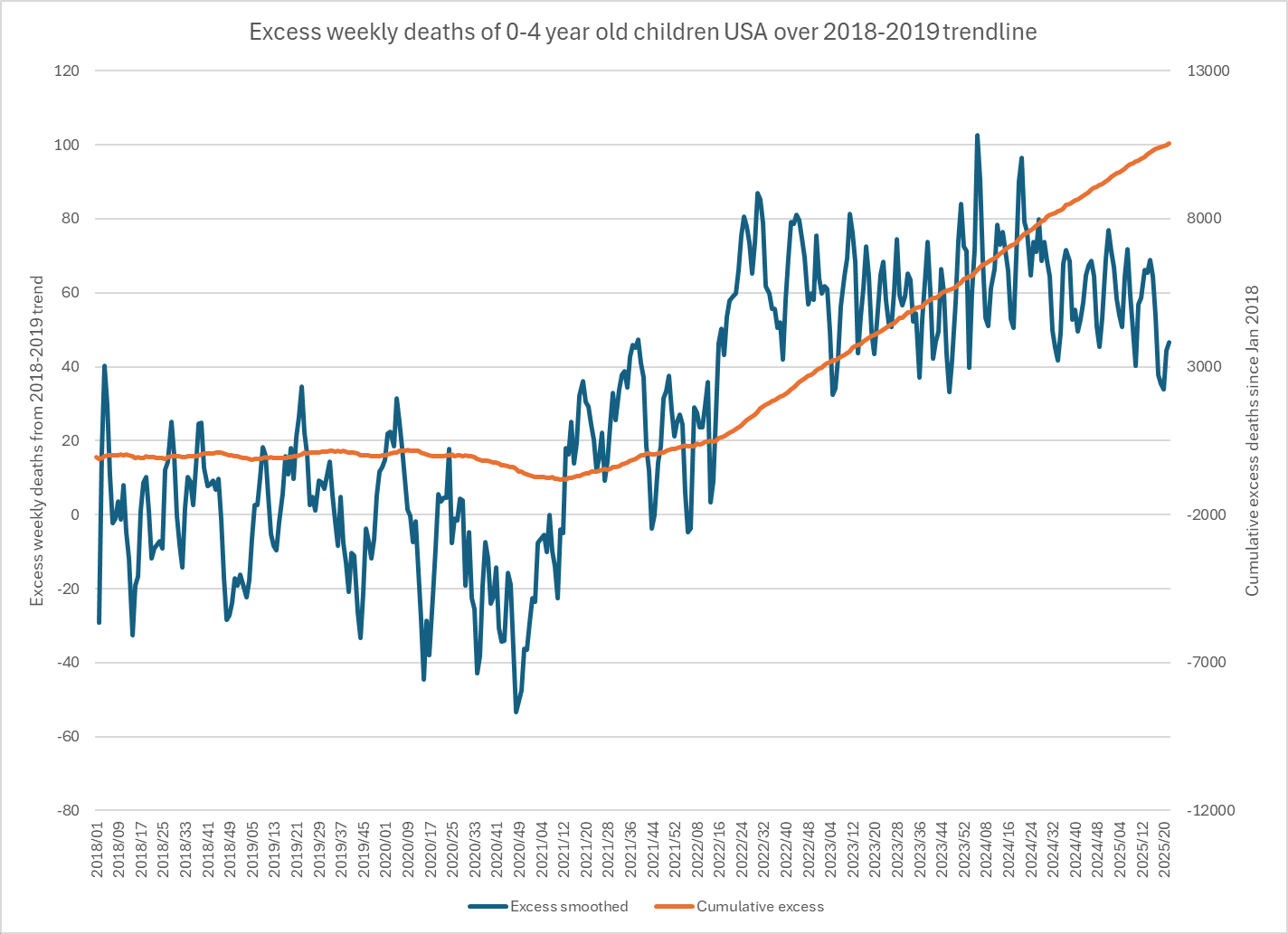
In recent years in the United States, over 80% of all deaths in ages 0-4 have occurred at age 0, so most of the deaths in Ethical Skeptic's plot are infant deaths. But the number of infant deaths is proportional to the number of births. The next plot shows that relative to a 2016-2019 linear trend, there were about -3% excess births in 2020 but there have been positive excess births since 2022, which partially explains why ages 0-4 had low deaths in 2020 but relatively high deaths since 2022:
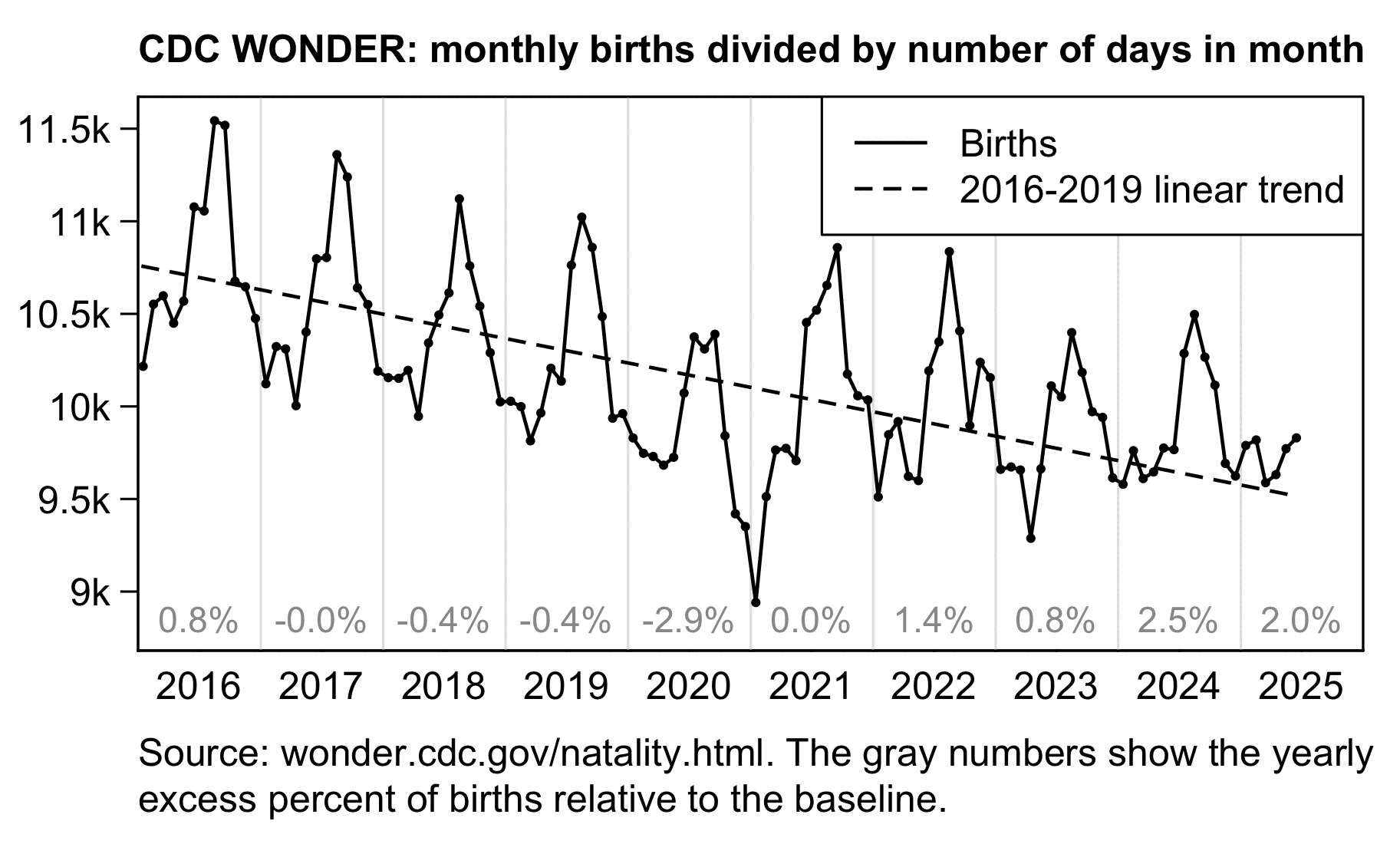
leg=c("Births","2018-2020 linear trend")
t=fread("https://sars2.net/f/wondernatality.csv")
t[,date:=as.Date(paste0(year,"-",month,"-16"))]
t[,born:=born/lubridate::days_in_month(date)]
xstart=as.Date("2016-1-1");xend=as.Date("2026-1-1")
xbreak=seq(xstart,xend,"6 month")
xlab=c(rbind(NA,2016:2025),NA)
t=t[t$date>=xstart&t$date<xend]
ylim=extendrange(c(t$born,t$trend),,c(.1,.05))
ybreak=pretty(t$born)
t$trend=predict(lm(born~date,t[year(date)%in%2016:2019]),t)
yearly=t[,sprintf("%.1f%%",(sum(born)/sum(trend)-1)*100),year]$V1
png("1.png",1825,1100,res=300)
par(mar=c(4.2,3,2.1,.8),mgp=c(0,.6,0),adj=0,lend="square")
tit="CDC WONDER: monthly births divided by number of days in month"
sub="Source: wonder.cdc.gov/natality.html. The gray numbers show the yearly
excess percent of births relative to the baseline."
leg=c("Births","2016-2019 linear trend")
plot(t$date,t$born,type="n",main=tit,xlab=NA,ylab=NA,xaxs="i",yaxs="i",yaxt="n",xaxt="n",ylim=ylim,xlim=c(xstart,xend),cex.main=1)
axis(1,at=xbreak,labels=xlab,tck=0,padj=-.6)
axis(2,at=ybreak,labels=paste0(ybreak/1e3,"k"),las=1,tck=-.03)
abline(v=seq(xstart,xend,"year"),col="gray90")
lines(t$date,t$born,lwd=1.5)
points(t$date,t$born,pch=20,cex=.6)
lines(t$date,t$trend,type="l",lty=2,lwd=1.5)
text(xbreak[c(F,T)],ylim[1],yearly,col="gray60",offset=.4,pos=3,cex=.93)
rect(xstart,ylim[1],xend,ylim[2])
mtext(text=sub,side=1,line=2.7,adj=0)
legend("topright",legend=leg,lty=c(1,2),lwd=1.5)
dev.off()
A 2018-2020 linear trend for births was even steeper, so it gave me about 8% excess births in 2024:
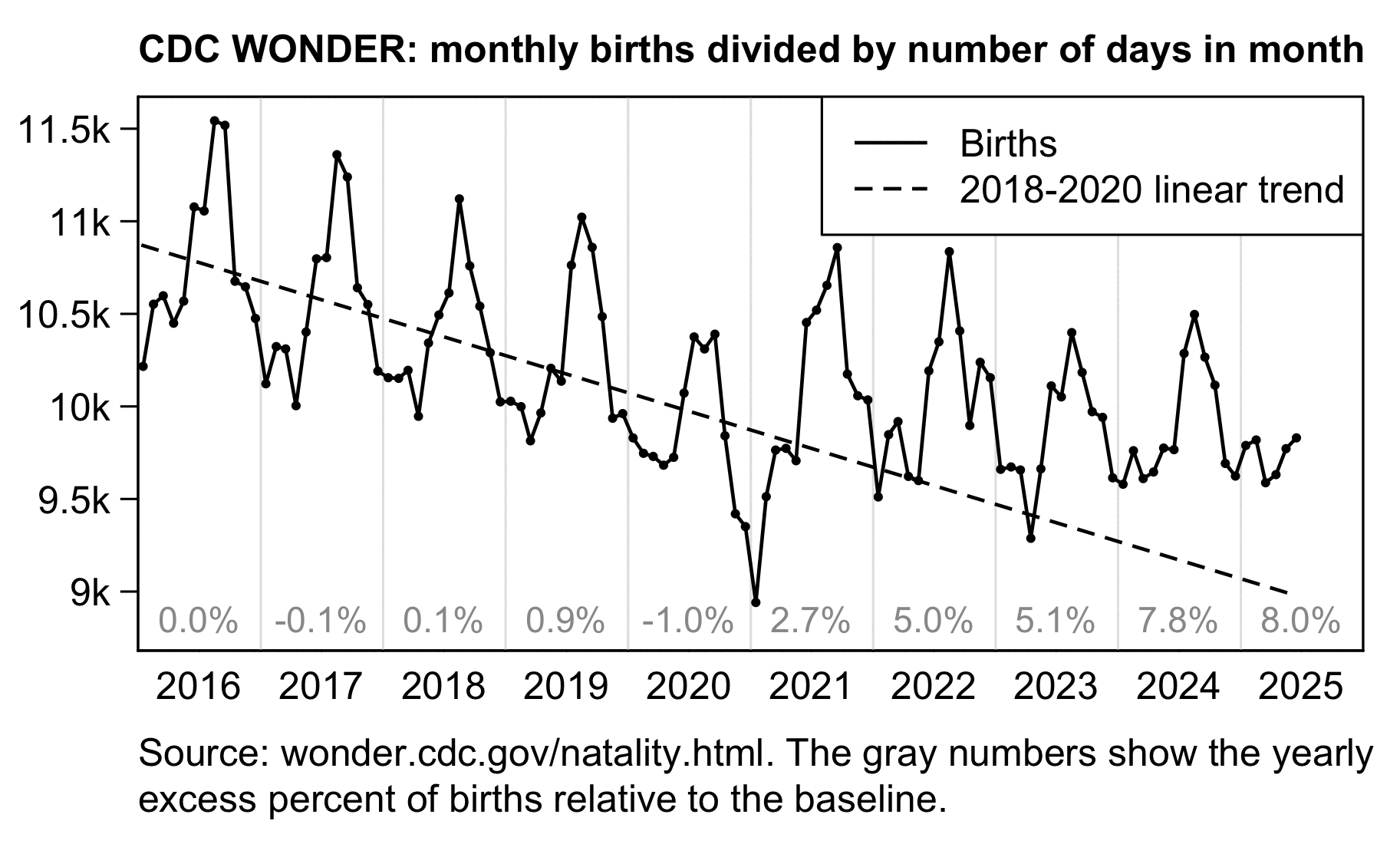
Ethical Skeptic's line labeled "emergent trend" also looks too steep, because I think he drew it by hand without doing any kind of a regression. The line starts in late 2020, which suggests that the line is supposed to approximate the trend in 2021-2025, or possibly from late 2020 until 2025. But the red line here shows that the real trend in 2021-2025 was much less steep, and even the trend in 2020-2025 was nowhere near as steep than Ethical Skeptic's "emergent trend":
t=fread("https://sars2.net/f/wondervaccinial0to4.csv")
t=t[!(year==2025&week>20)]
t[,x:=MMWRweek::MMWRweek2Date(year,week,4)]
p1=t[,.(x,y=dead,z=1,fit=T)]
p2=copy(p1)[,z:=2][,fit:=year(x)>2019]
p3=copy(p1)[,z:=3][,fit:=year(x)>2020]
p4=copy(p1)[,z:=4][,fit:=year(x)%in%2021:2024]
p5=copy(p1)[,z:=5][,fit:=year(x)%in%2022:2024]
p=rbind(p1,rbind(p2,p3,p4,p5)[,.(x,y=predict(lm(y~x,.SD[fit==T]),.SD),fit),z])
p[,z:=factor(z,,c("Weekly deaths","2020-2025 trend","2021-2025 trend","2021-2024 trend","2022-2024 trend"))]
xstart=as.Date("2017-7-1");xend=as.Date("2025-7-1");xbreak=seq(as.Date("2018-1-1"),xend-1,"6 month")
xlab=ifelse(month(xbreak)==7,year(xbreak),"")
ystart=250;yend=550;ybreak=0:11*50
ggplot(p)+
geom_point(aes(x,y,color=z,alpha=z),shape=3,stroke=.5,size=.8)+
geom_line(aes(x,y,color=z,linewidth=z),linetype="11")+
geom_line(data=p[fit==T],aes(x,y,color=z,linewidth=z))+
labs(title="Deaths in ages 0-4 with UCD A-R or 999--999",x=NULL,y=NULL)+
scale_x_continuous(limits=c(xstart,xend),breaks=xbreak,labels=xlab)+
scale_y_continuous(limits=c(ystart,yend),breaks=ybreak)+
scale_color_manual(values=c("black",hsv(5:6/6,.6,1),hsv(1:2/12,1,.7)))+
scale_linewidth_manual(values=c(0,.6,.6,.6,.6))+
scale_alpha_manual(values=c(1,0,0,0,0))+
guides(color=guide_legend(ncol=3,byrow=F))+
coord_cartesian(clip="off",expand=F)+
theme(axis.text=element_text(size=10,color="black"),
axis.ticks=element_line(color=alpha("black")),
axis.ticks.length=unit(4,"pt"),
axis.ticks.x=element_line(color=alpha("black",c(1,0))),
legend.background=element_blank(),
legend.box.spacing=unit(0,"pt"),
legend.key=element_blank(),
legend.key.height=unit(12,"pt"),
legend.key.width=unit(23,"pt"),
legend.margin=margin(-2,,14),
legend.position="top",
legend.spacing.x=unit(2,"pt"),
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=10,vjust=.5),
legend.title=element_blank(),
panel.background=element_blank(),
panel.grid=element_blank(),
plot.background=element_rect(fill="transparent",color=NA),
plot.title=element_text(size=11,hjust=.5))
ggsave("1.png",width=5,height=3.7,dpi=300*4)
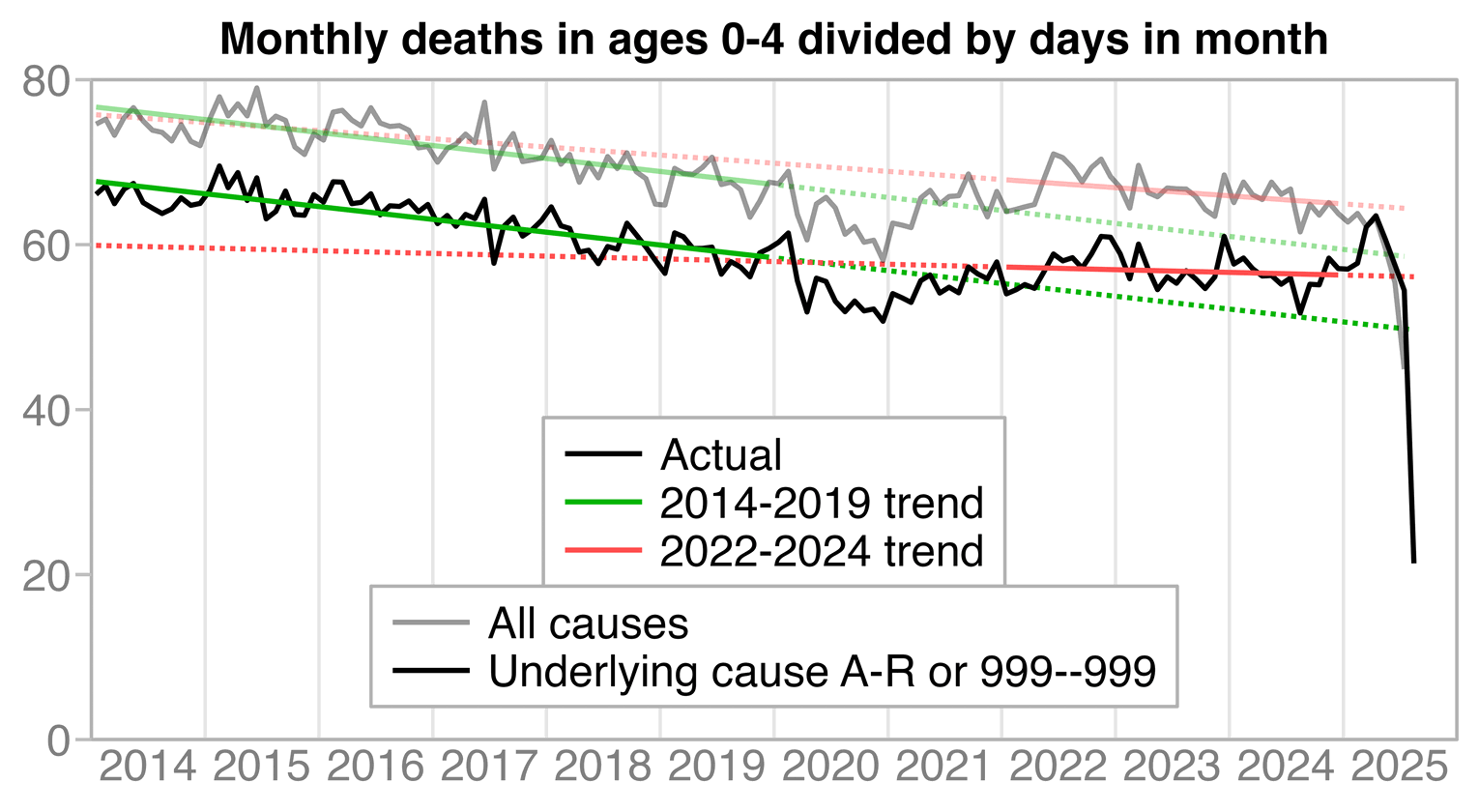
The deaths in 2025 are inflated by R96, R99, and 999--999 deaths. So it's probably better to exclude 2025 from the fitting period of the post-vaccine trend. The deaths were also unusually low in 2021. So 2022-2024 is the period I would choose for the post-vaccine trend, but the yellow line in the plot above shows that the 2022-2024 trend is sloped downwards.
The yearly total number of deaths also went down between 2022 and 2023, and between 2023 and 2024:
t=fread("https://sars2.net/f/wondervaccinial0to4ator999monthly.csv")
t[year%in%2018:2024&age<5,.(dead=sum(dead)),,year]
# year dead # UCD A-R or 999--999
# 2018 22073
# 2019 21403
# 2020 19928
# 2021 20176
# 2022 20944
# 2023 20758
# 2024 20560
The next plot shows how the slope of the trend in 2022-2024 is fairly similar to the trend in 2014-2019, even though I didn't even do any adjustment for population size or the number of births:
t1=fread("https://sars2.net/f/uspopdeadmonthly.csv")
t2=fread("https://sars2.net/f/wondervaccinial0to4ator999monthly.csv")
t=rbind(t1[,.(year,month,age,dead,z=1)],t2[,z:=2])[age<5]
p=t[,.(y=sum(dead)),.(x=as.Date(paste(year,month,16,sep="-")),z)]
p[,y:=y/days_in_month(x)]
p=na.omit(p)
p1=p[,type:=1][,fit:=T]
p2=copy(p)[,type:=2][,fit:=year(x)%in%2014:2019]
p3=copy(p)[,type:=3][,fit:=year(x)%in%2022:2024]
p=rbind(p,rbind(p2,p3)[,.(x,y=predict(lm(y~x,.SD[fit==T]),.SD),fit),.(z,type)])
p[,z:=factor(z,,c("All causes","Underlying cause A-R or 999--999"))]
p[,type:=factor(type,,c("Actual","2014-2019 trend","2022-2024 trend"))]
p=p[year(x)%in%2014:2025]
xstart=as.Date("2014-1-1");xend=as.Date("2026-1-1");xbreak=seq(xstart+182,xend,"year")
ybreak=pretty(c(0,p$y));ystart=0;yend=max(ybreak)
ggplot(p)+
geom_vline(xintercept=seq(xstart,xend,"year"),color="gray90",linewidth=.4)+
annotate("rect",xmin=xstart,xmax=xend,ymin=ystart,ymax=yend,linewidth=.4,lineend="square",fill=NA,color="gray70")+
geom_line(data=copy(p)[fit==T,y:=NA],aes(x,y,alpha=z,color=type),linewidth=.6,linetype="11",show.legend=F)+
geom_line(data=p[fit==T],aes(x,y,alpha=z,color=type),linewidth=.6)+
labs(x=NULL,y=NULL,title="Monthly deaths in ages 0-4 divided by days in month")+
scale_x_continuous(limits=c(xstart,xend),breaks=xbreak,labels=year(xbreak))+
scale_y_continuous(limits=c(ystart,yend),breaks=ybreak)+
scale_color_manual(values=c("black",hsv(1/3,1,.7),hsv(0,.7,1)))+
scale_alpha_manual(values=c(.4,1))+
coord_cartesian(clip="off",expand=F)+
guides(color=guide_legend(ncol=1,byrow=F))+
theme(axis.text=element_text(size=11,color="gray50"),
axis.text.y=element_text(margin=margin(,1)),
axis.ticks=element_line(linewidth=.4,color="gray75"),
axis.ticks.length=unit(0,"pt"),
axis.ticks.length.y=unit(4,"pt"),
legend.background=element_rect(color="gray70",linewidth=.4),
legend.box.just="center",
legend.box.spacing=unit(0,"pt"),
legend.direction="vertical",
legend.justification=c(.5,0),
legend.key=element_blank(),
legend.key.height=unit(12,"pt"),
legend.key.width=unit(24,"pt"),
legend.margin=margin(3,5,3,3),
legend.position=c(.5,.05),
legend.spacing.x=unit(2,"pt"),
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=11),
legend.title=element_blank(),
panel.background=element_blank(),
plot.title=element_text(size=11,hjust=.5,face=2,margin=margin(,,4)))
ggsave("1.png",width=5.1,height=2.8,dpi=300*4)
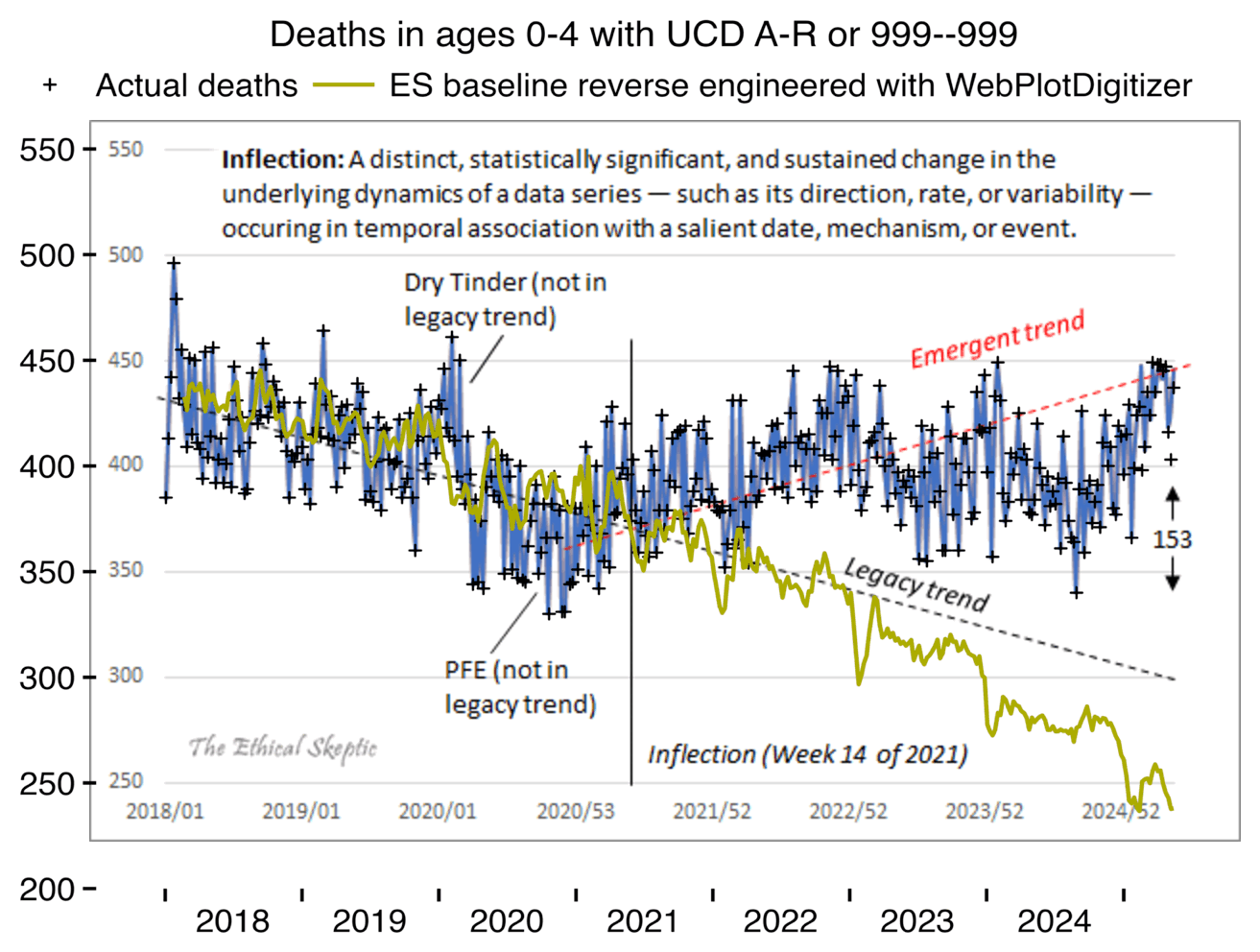
Even though Ethical Skeptic's legacy trend is extremely steep, his baseline I reverse engineered with WebPlotDigitizer is even steeper:
ma=\(x,b=1,f=b){x[]=rowMeans(embed(c(rep(NA,b),x,rep(NA,f)),f+b+1),na.rm=T);x}
t=fread("https://sars2.net/f/wondervaccinial0to4.csv")
t[,x:=MMWRweek::MMWRweek2Date(year,week,4)]
p=t[,.(year,week,x,y=c(dead,ma(dead,3,2)-excess),z=rep(1:2,each=.N))]
p=p[!(year==2025&week>20)]
p[,z:=factor(z,,c("Actual deaths","ES baseline reverse engineered with WebPlotDigitizer"))]
xstart=as.Date("2017-7-1");xend=as.Date("2025-7-1");xbreak=seq(as.Date("2018-1-1"),xend-1,"6 month")
xlab=ifelse(month(xbreak)==7,year(xbreak),"")
ystart=225;yend=550;ybreak=0:11*50
ggplot(p)+
geom_point(aes(x,y,color=z,alpha=z),shape=3,stroke=.5,size=.8)+
geom_line(aes(x,y,color=z,linewidth=z))+
labs(title="Deaths in ages 0-4 with UCD A-R or 999--999",x=NULL,y=NULL)+
scale_x_continuous(limits=c(xstart,xend),breaks=xbreak,labels=xlab)+
scale_y_continuous(limits=c(ystart,yend),breaks=ybreak)+
scale_color_manual(values=c("black","#aaaa00"))+
scale_linewidth_manual(values=c(0,.6,.6,.6,.6))+
scale_alpha_manual(values=c(1,0,0,0,0))+
guides(color=guide_legend(ncol=3,byrow=F))+
coord_cartesian(clip="off",expand=F)+
theme(axis.text=element_text(size=10,color="black"),
axis.ticks=element_line(color=alpha("black")),
axis.ticks.length=unit(4,"pt"),
axis.ticks.x=element_line(color=alpha("black",c(1,0))),
legend.background=element_blank(),
legend.box.spacing=unit(0,"pt"),
legend.key=element_blank(),
legend.key.height=unit(12,"pt"),
legend.key.width=unit(23,"pt"),
legend.margin=margin(-2,,14),
legend.position="top",
legend.spacing.x=unit(2,"pt"),
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=10,vjust=.5),
legend.title=element_blank(),
panel.background=element_blank(),
panel.grid=element_blank(),
plot.background=element_rect(fill="transparent",color=NA),
plot.title=element_text(size=11,hjust=.5))
ggsave("1.png",width=5,height=4,dpi=300*4)
ES should've also included a line similar to my yellow line in his plot, which would've shown the actual baseline he employed in his plot for weekly excess deaths. People on Twitter often complain to ES that his plots only show the excess deaths but not the baseline, so if he gets a massive increase in excess deaths, people can't tell if it's because the deaths are actually going up, or if he just calculated his baseline improperly.
Ethical Skeptic got 25 sigma excess deaths on weeks 14 to 20 of 2025 in his plot. The high sigma score is largely due to his baseline being too steep, deaths in 2025 being inflated by 999--999 and R99 deaths, and his 2-year baseline producing inflated z-scores.
ES calculates the sigma score by subtracting baseline deaths from actual deaths, and then dividing the result by the standard deviation of the residual deaths in the baseline fitting period (i.e. actual deaths minus baseline deaths in 2018-2019). If you fit a seasonality-adjusted baseline against only 2 years of data, it tends to produce a low residual number of deaths in the fitting period, because the baseline ends up adapting to whatever the deaths happen to be during the baseline period, so therefore you get a low standard deviation of the residuals and you subsequently get high sigma scores. If you fit a seasonality-adjusted baseline against only one year of data, you can get a standard deviation of zero for the residuals during the fitting period, and therefore you subsequently get infinite sigma scores. But similarly a 2-year baseline tends to produce lower residuals than a longer baseline.
In the next plot I did a linear regression of yearly data in 2010-2019, and I calculated the 95% prediction interval which shows the range where 95% of values are expected to fall assuming that the linear trend remains in place. The number of deaths in 2024 did not even cross above the 95% PI, even though the ASMR in 2024 did reach slightly above the PI. But the ASMR was still lower in 2024 than any year before 2020:
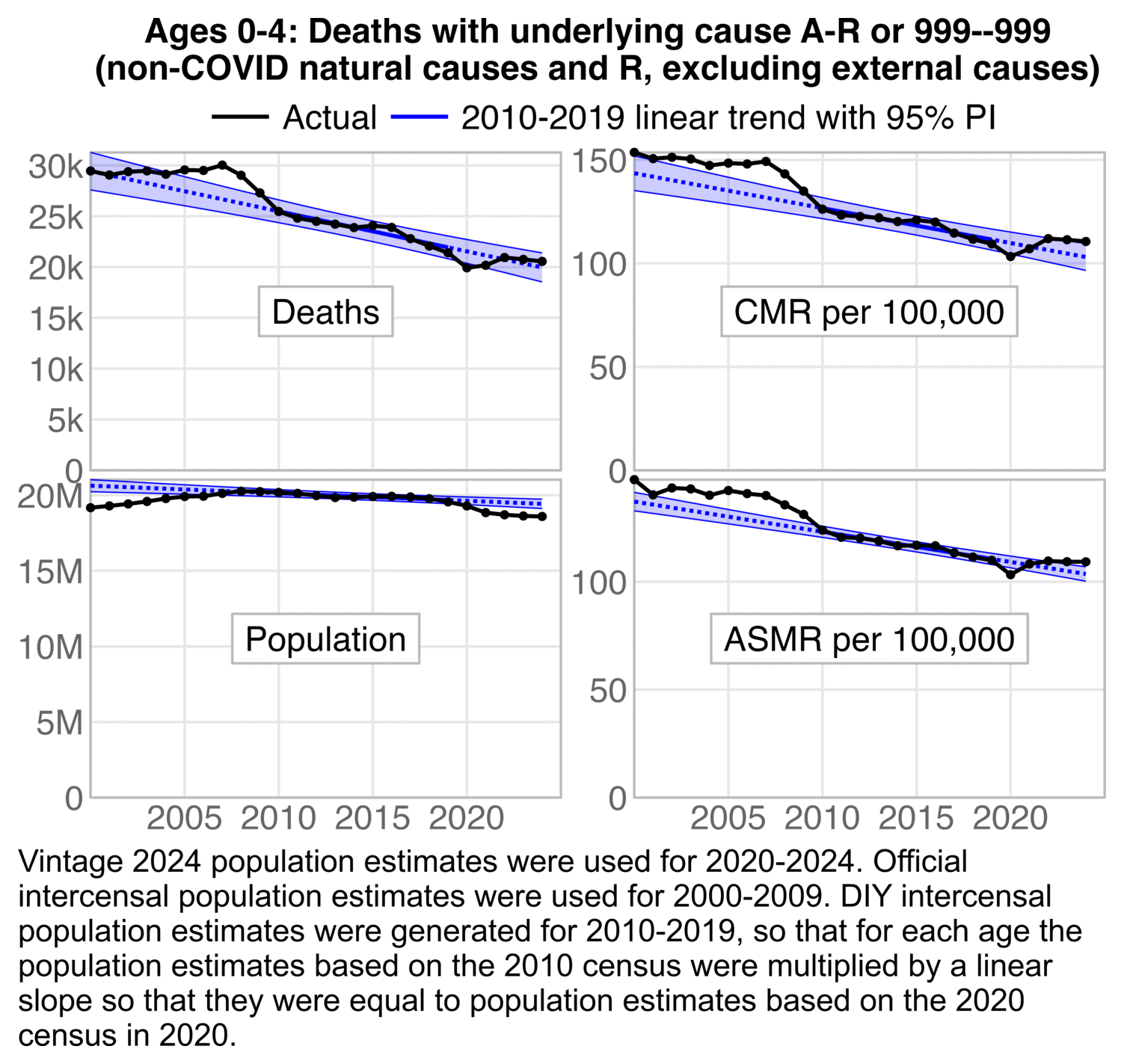
t=fread("https://sars2.net/f/wondervaccinial0to4ator999monthly.csv")
a=t[age<5&year%in%2000:2024,.(dead=sum(dead)),.(year,age)]
a=merge(a,fread("https://sars2.net/f/uspopdead.csv")[,.(age,year,pop)])
a=merge(a[year==2020,sum(pop),age][,.(age,std=V1/sum(V1))],a)
p=a[,.(y=c(sum(dead),sum(pop),sum(dead)/sum(pop)*1e5,sum(dead/pop*std*1e5)),facet=1:4),.(x=year)]
p=p[,cbind(.SD,predict(lm(y~x,.SD[x%in%2010:2019]),.SD,interval="prediction",level=.95)),facet]
p[,facet:=factor(facet,,c("Deaths","Population","CMR per 100,000","ASMR per 100,000"))]
lab=c("Actual","2010-2019 linear trend with 95% PI")
lab=factor(lab,unique(lab))
xstart=2000;xend=2025;xbreak=seq(2005,2020,5)
ybreak=pretty(c(0,p[,c(y,lwr,upr)]));ystart=0;yend=max(ybreak)
ylim=p[,.(max=max(y,upr)),facet]
ggplot(p)+
facet_wrap(~facet,ncol=2,dir="v",scales="free_y")+
geom_rect(data=ylim,aes(ymax=max),ymin=0,xmin=xstart,xmax=xend,lineend="square",linejoin="mitre",fill=NA,color="gray75",linewidth=.4)+
geom_ribbon(aes(x,ymin=lwr,ymax=upr),fill="blue",color="blue",alpha=.2,linewidth=.2)+
geom_line(aes(x,y,color=lab[1]),linewidth=.6)+
geom_line(aes(x,fit,color=lab[2]),linewidth=.6,show.legend=F,linetype="11")+
geom_line(data=p[x%in%2010:2019],aes(x,fit,color=lab[2]),linewidth=.6)+
geom_line(aes(x,y,color=lab[1]),linewidth=.6)+
geom_point(aes(x,y,color=lab[1]),stroke=0,size=1.4,show.legend=F)+
geom_label(data=ylim,aes(mean(c(xstart,xend)),max/2,label=facet),size=3.87,label.r=unit(0,"pt"),label.padding=unit(4,"pt"),label.size=.4,fill="white",color="gray75")+
geom_label(data=ylim,aes(mean(c(xstart,xend)),max/2,label=facet),size=3.87,label.r=unit(0,"pt"),label.padding=unit(4,"pt"),label.size=0,fill=NA,)+
labs(title="Ages 0-4: Deaths with underlying cause A-R or 999--999\n(non-COVID natural causes and R, excluding external causes)",x=NULL,y=NULL)+
scale_x_continuous(limits=c(xstart,xend),breaks=xbreak)+
scale_y_continuous(limits=\(x)c(0,max(x)),breaks=\(x)pretty(c(0,x)),labels=\(x)ifelse(x>=1e3,ifelse(x>=1e6,paste0(x/1e6,"M"),paste0(x/1e3,"k")),x))+
scale_color_manual(labels=lab,values=c("black","blue"))+
coord_cartesian(clip="off",expand=F)+
theme(axis.text=element_text(size=11,color="gray40",margin=margin(8,8,8,8)),
axis.ticks.length=unit(0,"pt"),
legend.background=element_blank(),
legend.box.spacing=unit(0,"pt"),
legend.direction="horizontal",
legend.key=element_blank(),
legend.key.height=unit(11,"pt"),
legend.key.width=unit(23,"pt"),
legend.margin=margin(,,6),
legend.position="top",
legend.spacing.x=unit(2,"pt"),
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=11),
legend.title=element_blank(),
panel.background=element_blank(),
panel.grid.major=element_line(linewidth=.4,color="gray90"),
panel.spacing=unit(3,"pt"),
plot.title=element_text(size=11,hjust=.5,face="bold",margin=margin(,,4)),
strip.background=element_blank(),
strip.text=element_blank())
ggsave("1.png",width=5,height=3.8,dpi=300*4)
The title of the plot that shows excess deaths in ages 0-4 says "Mortality Deviation From 2018/19 Trend":

In this tweet ES wrote that the baseline in his plot was fitted against week 1 of 2018 to week 50 of 2019: [https://x.com/EthicalSkeptic/status/1950916264928358512]
The methodology image in his blog post also says that "Legacy years 2018-2019 serve as stable pre-Covid benchmarks": [https://theethicalskeptic.com/wp-content/uploads/2025/08/Methods-and-Data-Considerations.webp]

But on the other hand in the caption for the plot below, ES wrote that his "analysis does not rely solely upon 2018/19 as is claimed by dishonest parties". I don't know if he referred only to the plot below, where the "legacy trend" appears to approximate the trend in 2018 to 2020, or if he also referred to the plot for the excess deaths. By "dishonest parties" he might have referred to me, but I never claimed that the plot below had a 2018-2019 baseline, but only that the plot for excess deaths was supposed to have a 2018-2019 baseline: [https://theethicalskeptic.com/2025/08/19/houston-we-have-another-problem/]
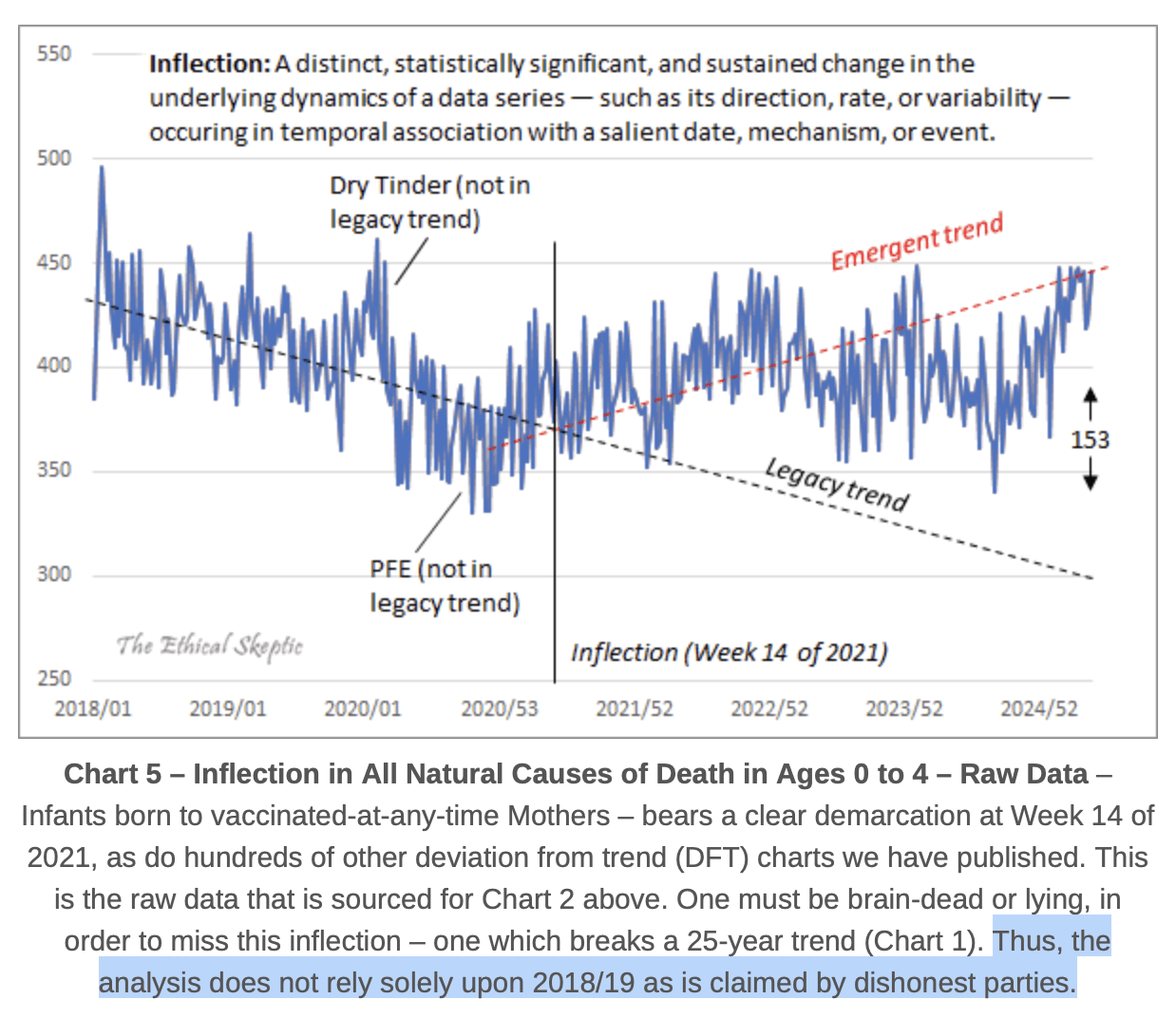
In his blog post ES also wrote: "The brief spike visible in late 2019 and early 2020 reflects a dry-tinder effect - mortality rising among already-vulnerable populations - occurring during a period when the virus was either not yet formally detected or not yet recognized as having reached U.S. shores. This is followed by a subtle pull-forward effect (PFE) visible in the variance data over the subsequent eight months. Neither of these 2020 artifacts is incorporated into the baseline trend alignment of the chart, which, as the reader will note, remains anchored to the more stable 2018-2019 period." [https://theethicalskeptic.com/2025/08/19/houston-we-have-another-problem/]
But if he did not consider data from 2020 when he calculated his baseline, then why does his baseline take such a steep turn downwards between 2019 and 2020?
ES might have deliberately falsified his baseline, because I have found him performing similar undocumented adjustments in his other plots. When I used WebPlotDigitizer to reverse engineer the baseline in his plot for cancer deaths in ages 0-54, his baseline had a similar unexplained downward turn between 2019 and 2020, even though ES says he didn't apply PFE adjustment to the plot: [ethical.html#Deaths_from_malignant_neoplasms_in_ages_0_to_54]
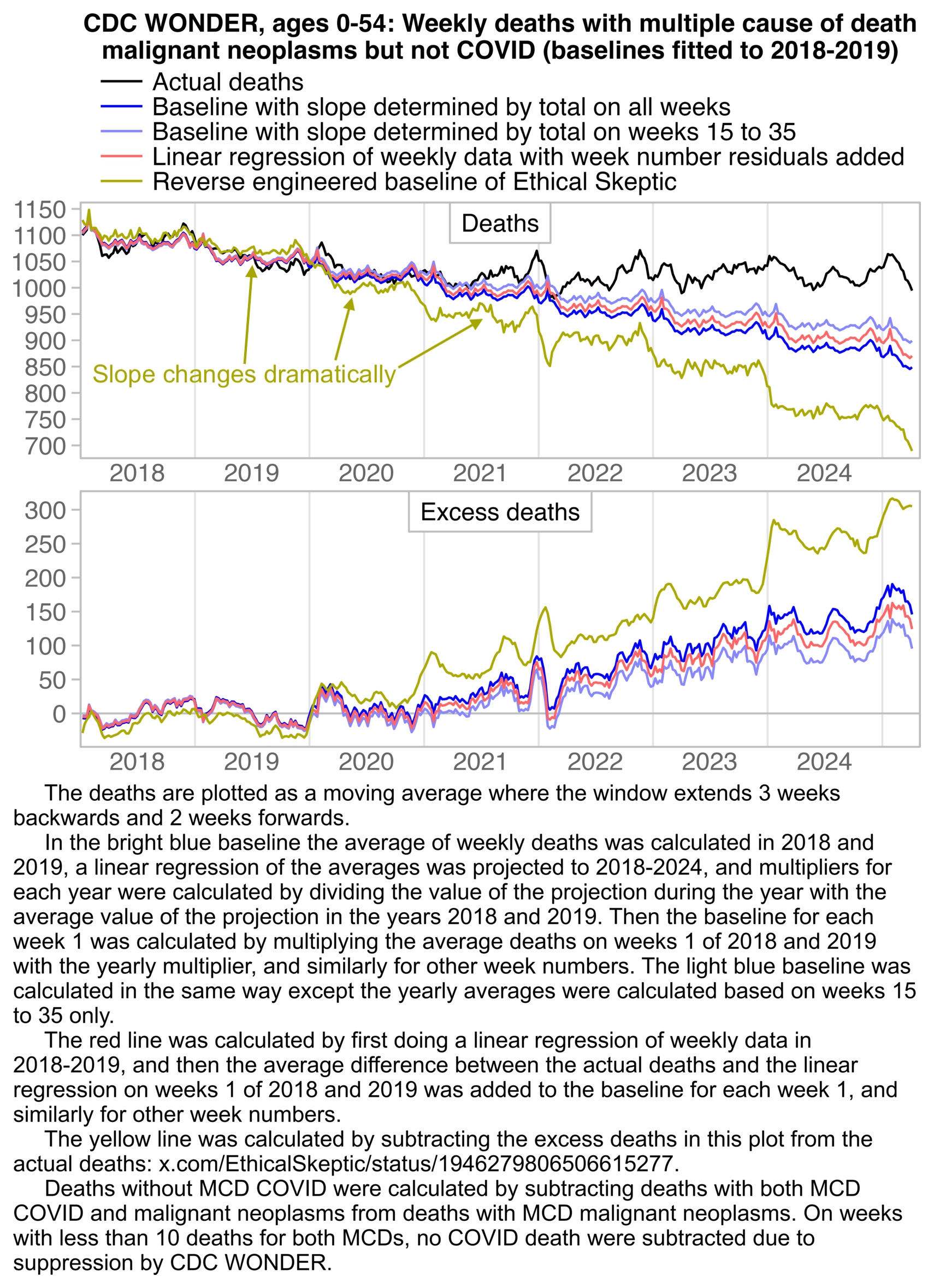
The baseline in the plot above also has a similar step pattern as the baseline in the vaccinial plot, where there's a sudden drop in the baseline between late 2023 and early 2024, and between late 2024 and early 2025.
ES included this plot from a CDC report in his blog post: [https://theethicalskeptic.com/2025/08/19/houston-we-have-another-problem/, https://www.cdc.gov/nchs/data/nvsr/nvsr74/nvsr74-07.pdf]
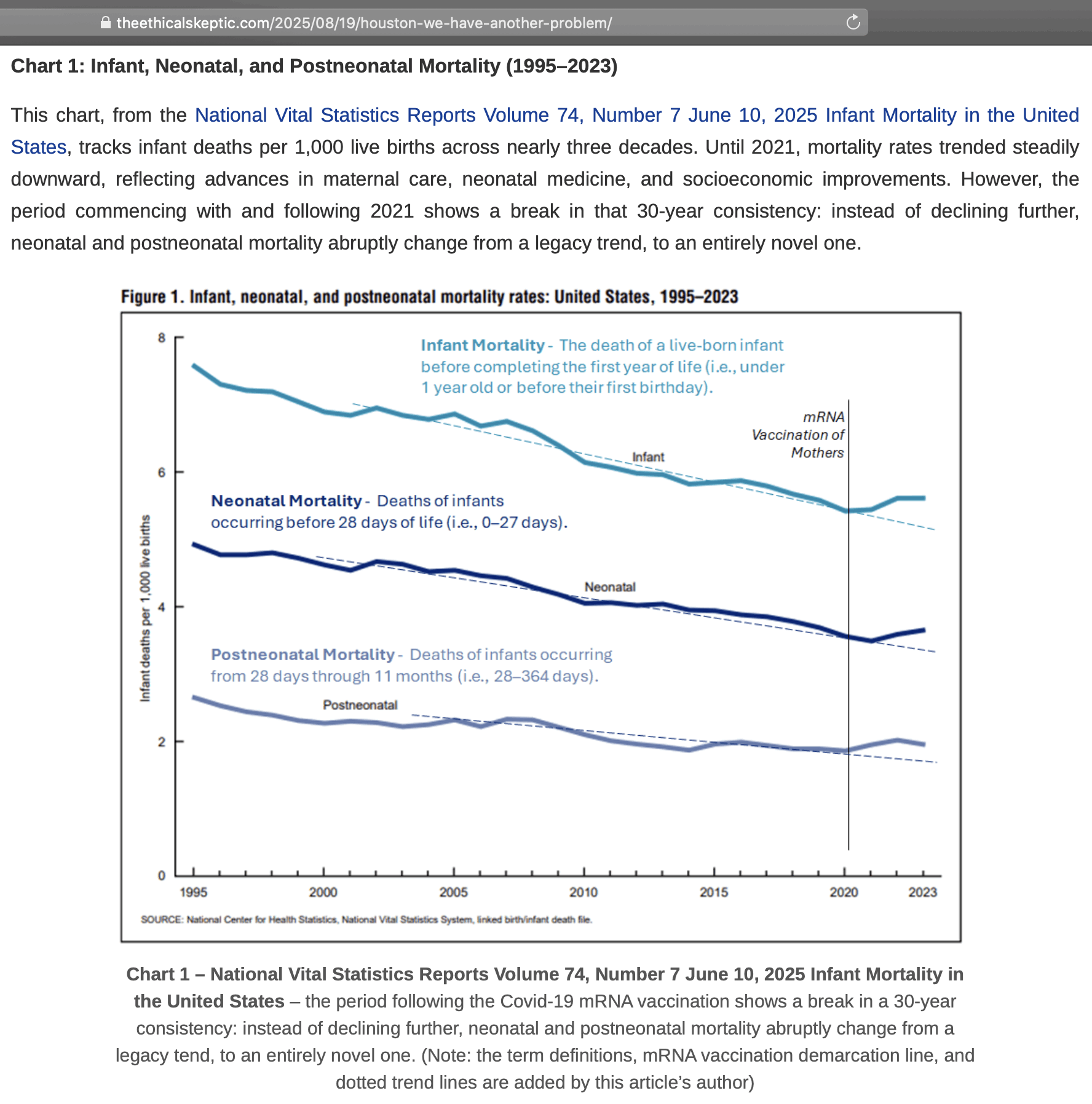
The plot was missing 2024, but the next plot shows that the ratio of infant deaths per thousand live births was slightly lower in 2024 than 2023. There was a decreasing trend in infant mortality in the 2010s which seems to have flattened out in the 2020s, but it's not clear if the flattening out has anything to do with COVID vaccines, and there was previously a similar flat period around the years 2000 to 2005: [https://wonder.cdc.gov/natality.html]
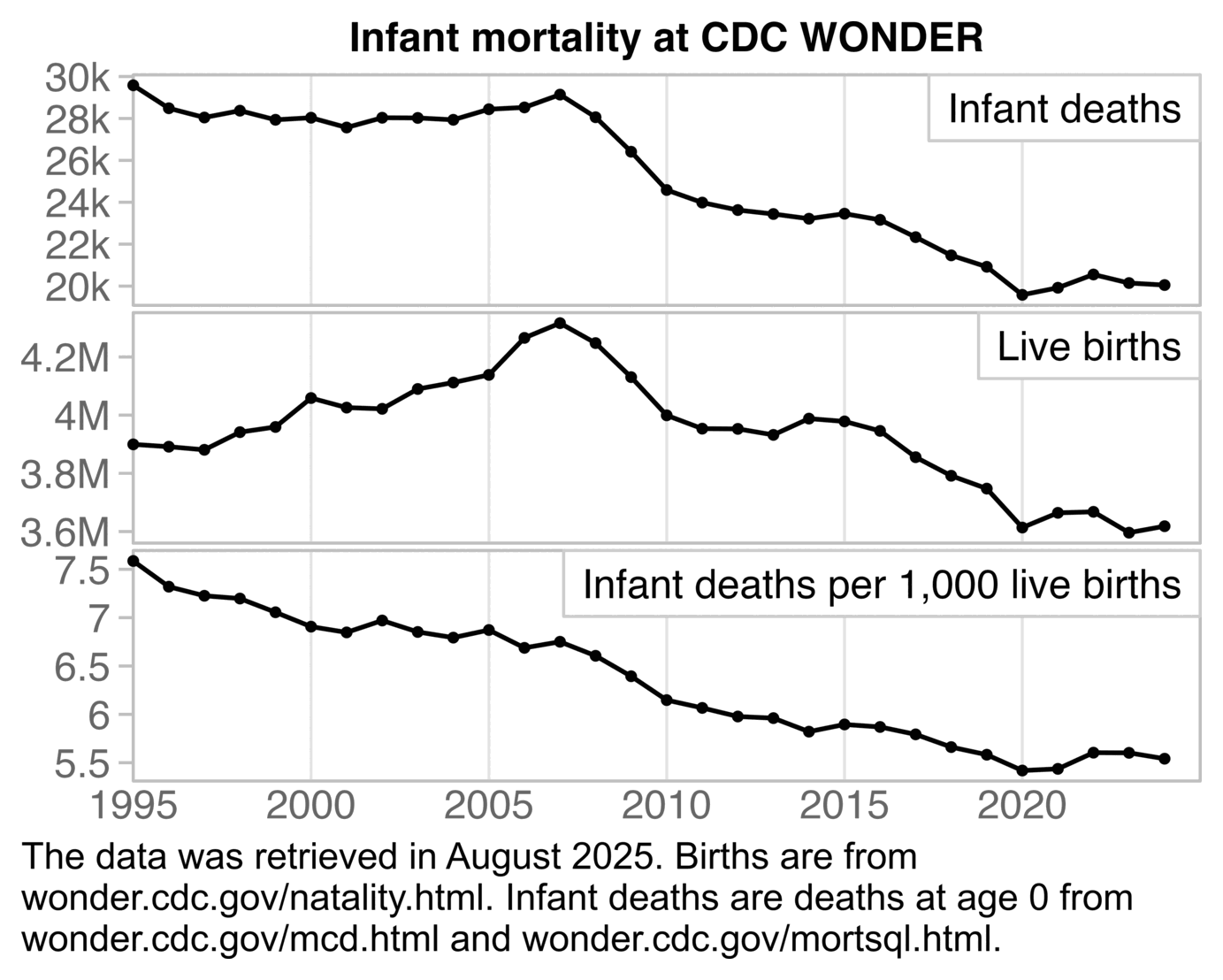
t=fread("https://sars2.net/f/wondernatality.csv")
t=t[,.(born=sum(born)),year]
t=rbind(t,list(1995:2002,c(3899589,3891494,3880894,3941553,3959417,4058814,4025933,4021726)))
d=fread("https://sars2.net/f/uspopdead.csv")[age==0,.(year,dead)]
d=rbind(d,list(1995:1998,c(29583,28487,28045,28371)))
p=merge(t,d)[year<2025,.(x=year,y=c(dead,born,dead/born*1e3),z=rep(1:3,each=.N))]
p[,z:=factor(z,,c("Infant deaths","Live births","Infant deaths per thousand live births"))]
xstart=1995;xend=2024;xbreak=seq(xstart,xend,5)
p=p[x%in%xstart:xend]
ylim=p[,{x=extendrange(y);.(min=x[1],max=x[2])},z]
ggplot(p)+
facet_wrap(~z,ncol=1,dir="v",scales="free")+
geom_rect(data=ylim,aes(ymin=min,ymax=max),xmin=xstart-.5,xmax=xend+.5,lineend="square",linejoin="mitre",fill=NA,color="gray75",linewidth=.4)+
geom_line(aes(x,y),linewidth=.6)+
geom_point(aes(x,y),stroke=0,size=1.5)+
geom_label(data=ylim,aes(xend+.5,max,label=z),vjust=1,hjust=1,label.r=unit(0,"pt"),label.padding=unit(5,"pt"),label.size=.4,size=3.87,fill="white",color="gray80")+
geom_label(data=ylim,aes(xend+.5,max,label=z),vjust=1,hjust=1,label.r=unit(0,"pt"),label.padding=unit(5,"pt"),label.size=0,size=3.87,fill=NA)+
labs(title="Infant mortality at CDC WONDER",x=NULL,y=NULL)+
scale_x_continuous(limits=c(xstart-.5,xend+.5),breaks=xbreak)+
scale_y_continuous(breaks=pretty,labels=\(x)ifelse(x>=1e3,paste0(x/1e3,"k"),x))+
scale_alpha_manual(values=c(1,0))+
coord_cartesian(clip="off",expand=F)+
theme(axis.text=element_text(size=11,color="gray40",margin=margin(2,2,2,2)),
axis.ticks=element_line(linewidth=.4,color="gray75"),
axis.ticks.length.x=unit(0,"pt"),
axis.ticks.length.y=unit(4,"pt"),
panel.background=element_blank(),
panel.grid=element_blank(),
panel.spacing=unit(2,"pt"),
plot.title=element_text(size=11,hjust=.5,face="bold",margin=margin(1,,4)),
strip.background=element_blank(),
strip.text=element_blank())
ggsave("1.png",width=4.6,height=4,dpi=300*4)
ES pointed out that the CDC report only had a 3% increase in infant mortality between 2021 and 2022: [https://x.com/EthicalSkeptic/status/1964730950660845778]
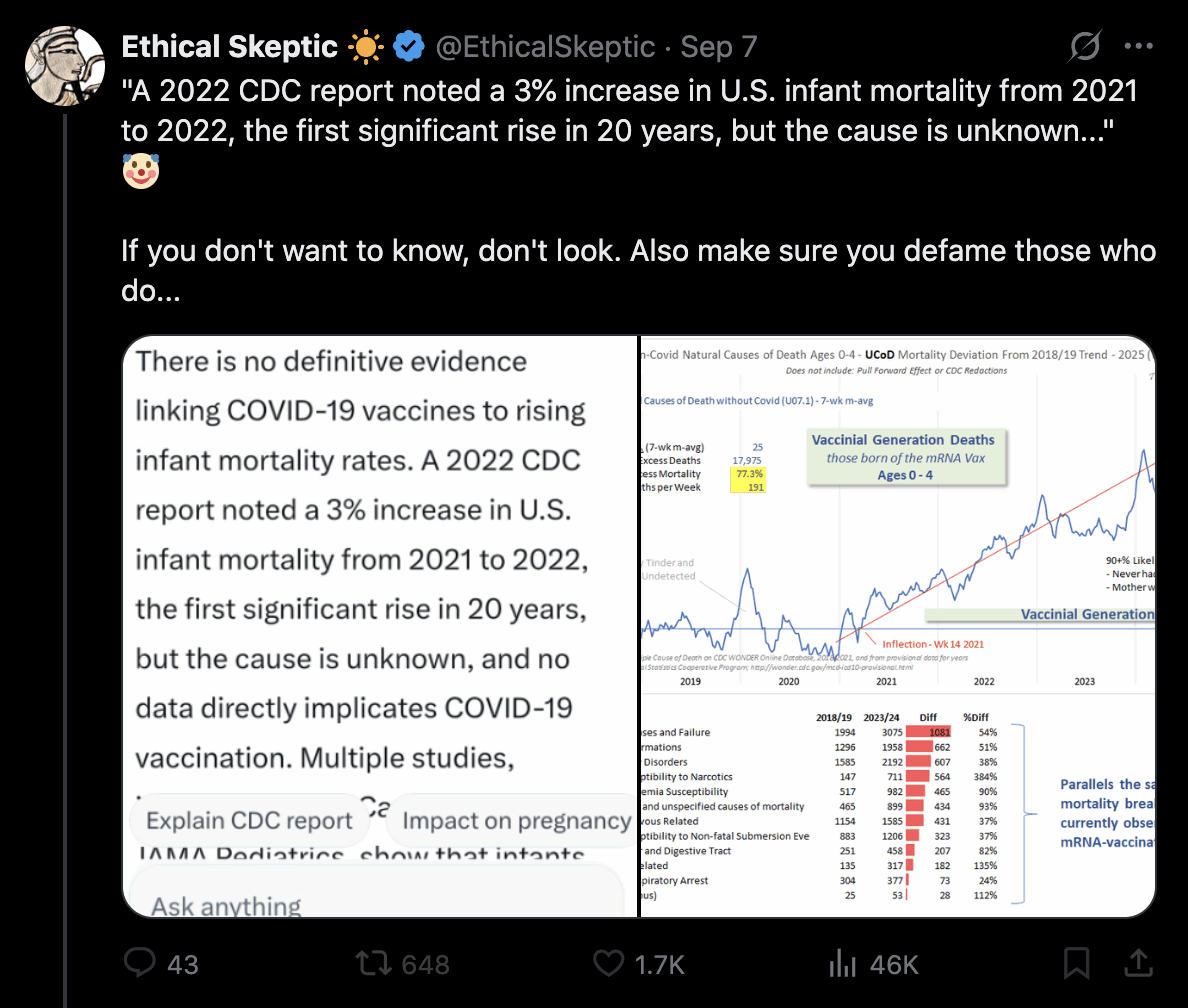
The CDC report specifically showed that the infant mortality rate per 1,000 live births was about 5.44 in 2021, 5.61 in 2022, and 5.61 in 2023. [https://www.cdc.gov/nchs/data/nvsr/nvsr74/nvsr74-07.pdf#page=8]
About 89% of the deaths in Ethical Skeptic's plot for ages 0-4 are deaths at age 0, so the results of his plot shouldn't look too different from a plot that only includes age 0. But based on my reverse engineered baseline for his plot, the yearly average of his weekly excess deaths increased from about 4% in 2021 to 15% in 2022, and further to about 26% in 2023:
t=fread("https://sars2.net/f/wondervaccinial0to4.csv")
a=na.omit(t)[,.(dead=mean(dead),excess=mean(excess)),year]
round(a[,.(year,dead,base=dead-excess)][,excesspct:=(dead/base-1)*100])
# year dead base excesspct
# 2018 419 428 -2
# 2019 410 417 -2
# 2020 382 389 -2
# 2021 387 373 4
# 2022 402 348 15
# 2023 398 315 26
# 2024 393 278 41
# 2025 423 248 70
But in comparison when I used a 1995-2019 linear regression as the baseline, the infant mortality rate in the CDC report was only about 5% above the baseline in 2022 and 6% in 2023:
t=CJ(year=1995:2023) t$rate=c(757,730,721,719,704,689,684,695,684,678,686,668,675,661,639,614,607,598,596,582,590,587,579,567,558,542,544,561,561)/100 t$base=predict(lm(rate~year,t[year<2020]),t) t[year>=2010,.(year,rate,base=round(base,2),excesspct=round((rate/base-1)*100))] # year rate base excesspct # 2010 6.14 6.27 -2 # 2011 6.07 6.20 -2 # 2012 5.98 6.12 -2 # 2013 5.96 6.04 -1 # 2014 5.82 5.97 -2 # 2015 5.90 5.89 0 # 2016 5.87 5.81 1 # 2017 5.79 5.73 1 # 2018 5.67 5.66 0 # 2019 5.58 5.58 0 # 2020 5.42 5.50 -1 # 2021 5.44 5.42 0 # 2022 5.61 5.35 5 # 2023 5.61 5.27 6
Here's a visual comparison of the excess mortality percentages:
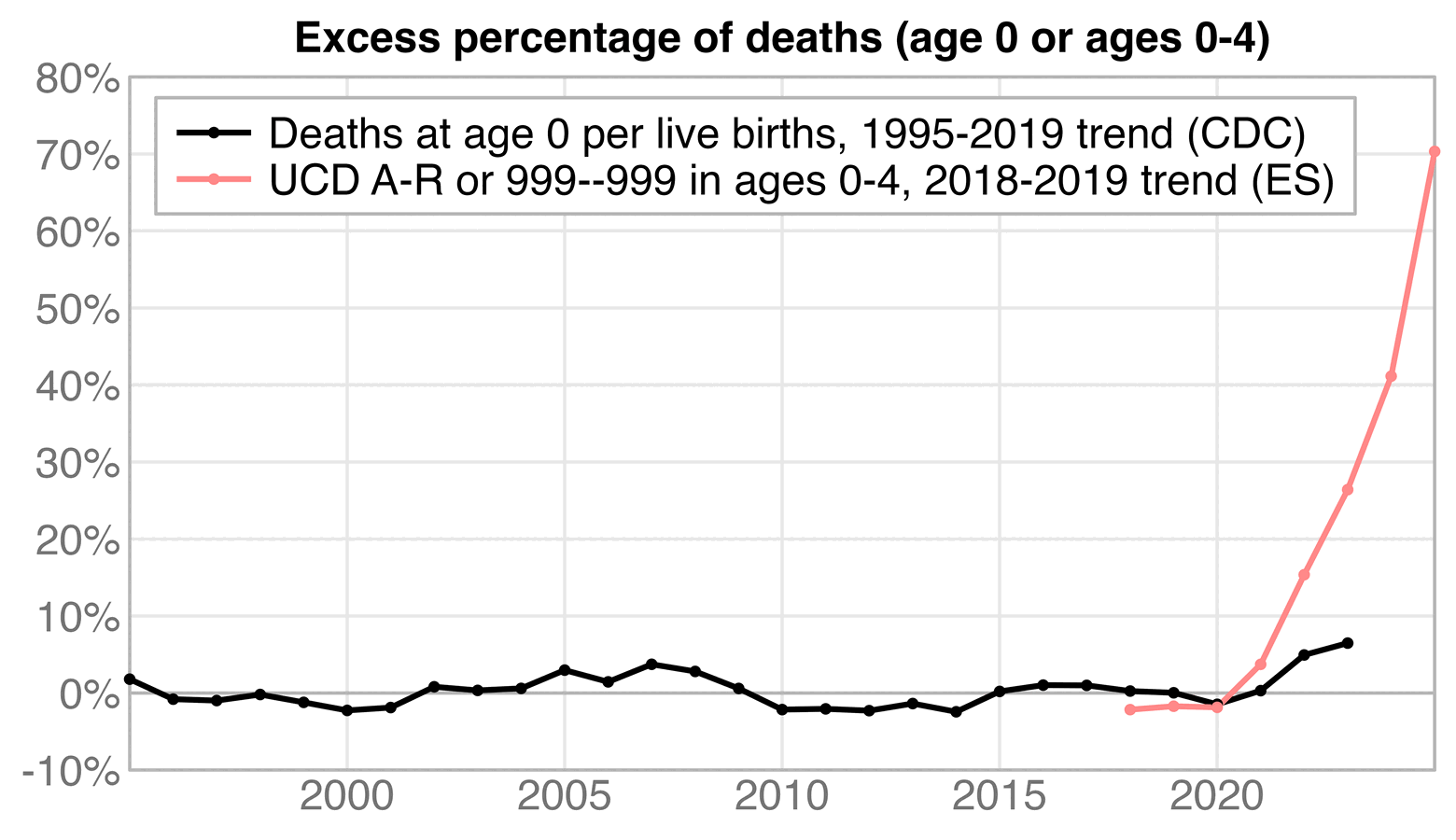
d=CJ(year=1995:2023)
d$rate=c(7.57,7.30,7.21,7.19,7.04,6.89,6.84,6.95,6.84,6.78,6.86,6.68,6.75,6.61,6.39,6.14,6.07,5.98,5.96,5.82,5.90,5.87,5.79,5.67,5.58,5.42,5.44,5.61,5.61)
d$base=predict(lm(rate~year,d[year<2020]),d)
p1=d[,.(x=year,y=(rate/base-1)*100,z=1)]
t=fread("https://sars2.net/f/wondervaccinial0to4.csv")
a=na.omit(t)[,.(dead=mean(dead),excess=mean(excess)),year]
p2=a[,.(x=year,y=(dead/(dead-excess)-1)*100,z=2)]
p=rbind(p1,p2)
p[,z:=factor(z,,c("Deaths at age 0 per live births, 1995-2019 trend (CDC)","UCD A-R or 999--999 in ages 0-4, 2018-2019 trend (ES)"))]
xmin=1995;xmax=2025;xbreak=seq(xmin+5,xmax-5,5)
ybreak=pretty(c(p$y),8);ymin=ybreak[1];ymax=max(ybreak)
ggplot(p)+
geom_hline(yintercept=0,color="gray70",linewidth=.4)+
annotate("rect",xmin=xmin,xmax=xmax,ymin=ymin,ymax=ymax,linewidth=.4,lineend="square",fill=NA,color="gray70")+
geom_line(aes(x,y,color=z),linewidth=.7)+
geom_point(aes(x,y,color=z),size=1.4,stroke=0)+
labs(x=NULL,y=NULL,title="Excess percentage of deaths (ages 0 or ages 0-4)")+
scale_x_continuous(limits=c(xmin,xmax),breaks=xbreak)+
scale_y_continuous(limits=c(ymin,ymax),breaks=ybreak,labels=\(x)paste0(x,"%"))+
scale_color_manual(values=c("black","#ff8888"))+
coord_cartesian(clip="off",expand=F)+
theme(axis.text=element_text(size=11,color="gray45"),
axis.ticks=element_line(linewidth=.4,color="black"),
axis.ticks.length=unit(0,"pt"),
legend.background=element_rect(color="gray70",linewidth=.4),
legend.box.spacing=unit(0,"pt"),
legend.justification=c(0,1),
legend.key=element_blank(),
legend.key.height=unit(12,"pt"),
legend.key.width=unit(24,"pt"),
legend.margin=margin(3,5,3,3),
legend.position=c(.02,.97),
legend.spacing.x=unit(2,"pt"),
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=11),
legend.title=element_blank(),
panel.background=element_blank(),
panel.grid.major=element_line(linewidth=.4,color="gray90"),
plot.title=element_text(size=11,hjust=.5,face=2,margin=margin(,,4)))
ggsave("1.png",width=5.2,height=3,dpi=300*4)
The 1995-2019 linear baseline might be too low by the 2020s relative to a longer-term curved trend. The CDC data started from 1995, so you couldn't see that the slope of the infant mortality trend changed drastically between the early 90s and late 90s. The next plot shows that there was relatively low infant mortality during the years at the start of the x-axis in the CDC plot, because each year between 1995 and 2001 was below the 1968-2019 exponential trend in infant mortality rate:
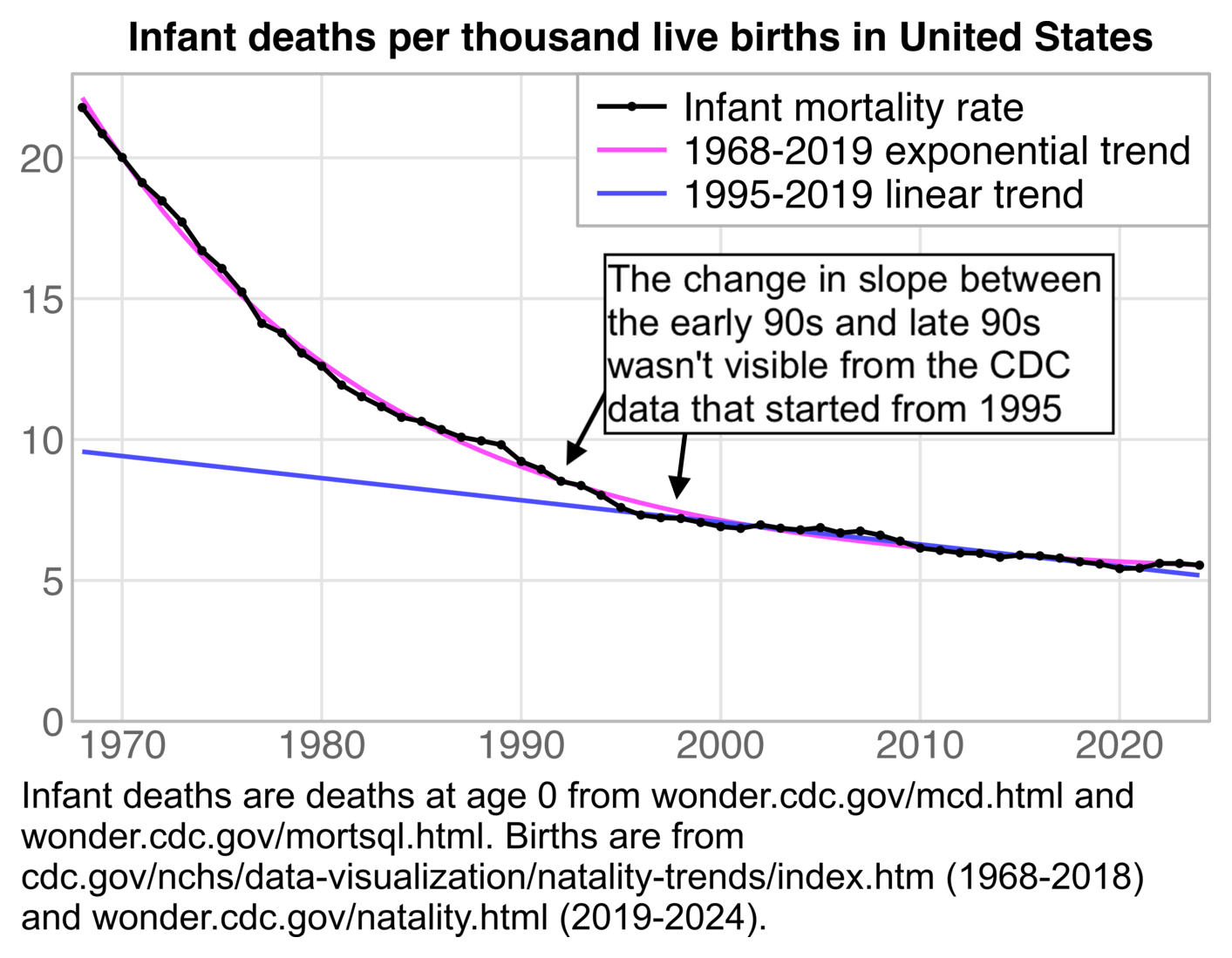
t=fread("https://sars2.net/f/NCHS_-_Births_and_General_Fertility_Rates__United_States.csv")
t=t[,.(year=Year,born=`Birth Number`)]
t=rbind(t,fread("https://sars2.net/f/wondernatality.csv")[year%in%2019:2024,.(born=sum(born)),year])
t=merge(fread("https://sars2.net/f/wonderinfantdeadpop.csv")[,.(year,dead)],t)
p1=t[,.(x=year,y=dead/born*1e3,z=1)]
p2=p1[,.(x,y=predict(nls(y~c+a*exp(b*(x-x[1])),.SD[x<2020],.SD[x<2020,.(a=y[.N]-y[1],b=-.08,c=min(y))]),.SD),z=2)]
p3=p1[,.(x,y=predict(lm(y~x,.SD[x%in%1995:2019]),.SD),z=3)]
p=rbind(p1,p2,p3)
p[,z:=factor(z,,c("Infant mortality rate","1968-2019 exponential trend","1995-2019 linear trend"))]
xstart=1968-.5;xend=2024+.5;xbreak=seq(1970,xend,10)
ylim=p[,{x=extendrange(y);.(min=0,max=x[2])}]
ggplot(p)+
geom_rect(data=ylim,aes(ymin=min,ymax=max),xmin=xstart,xmax=xend,lineend="square",linejoin="mitre",fill=NA,color="gray72",linewidth=.4)+
geom_line(aes(x,y,color=z),linewidth=.6)+
geom_point(aes(x,y,color=z,alpha=z),stroke=0,size=1.2)+
geom_line(data=p[z==z[1]],aes(x,y,color=z),linewidth=.6)+
labs(title="Infant deaths per thousand live births in United States",x=NULL,y=NULL)+
scale_x_continuous(limits=c(xstart,xend),breaks=xbreak)+
scale_color_manual(values=c("black",hsv(5:3/6,.7,1)))+
scale_alpha_manual(values=c(1,0,0,0))+
coord_cartesian(clip="off",expand=F)+
theme(axis.text=element_text(size=11,color="gray40",margin=margin(2,2,2,2)),
axis.ticks.length=unit(0,"pt"),
legend.background=element_rect(color="gray70",linewidth=.4),
legend.box.spacing=unit(0,"pt"),
legend.direction="vertical",
legend.justification=c(1,1),
legend.key=element_blank(),
legend.key.height=unit(12,"pt"),
legend.key.width=unit(24,"pt"),
legend.margin=margin(3,5,3,3),
legend.position=c(1,1),
legend.spacing.x=unit(2,"pt"),
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=11,vjust=.5),
legend.title=element_blank(),
panel.background=element_blank(),
panel.grid=element_blank(),
panel.grid.major=element_line(linewidth=.4,color="gray90"),
plot.subtitle=element_text(hjust=.5,margin=margin(,,4)),
plot.title=element_text(size=11,hjust=.5,face="bold",margin=margin(,,4)))
ggsave("1.png",width=4.7,height=3,dpi=300*4)
About two thirds of deaths at age 0 in the United States are neonatal deaths, which refers to deaths during the first 28 days from birth. But contrary to Ethical Skeptic's hypothesis that an increase in infant deaths was caused by prenatal exposure to vaccines, in the next plot I got negative excess neonatal mortality from March 2021 up to April 2022, when many newborns had been exposed to COVID vaccines during pregnancy. But I got high excess neonatal mortality in 2023 and 2024, when few newborns had been exposed to COVID vaccines during pregnancy:
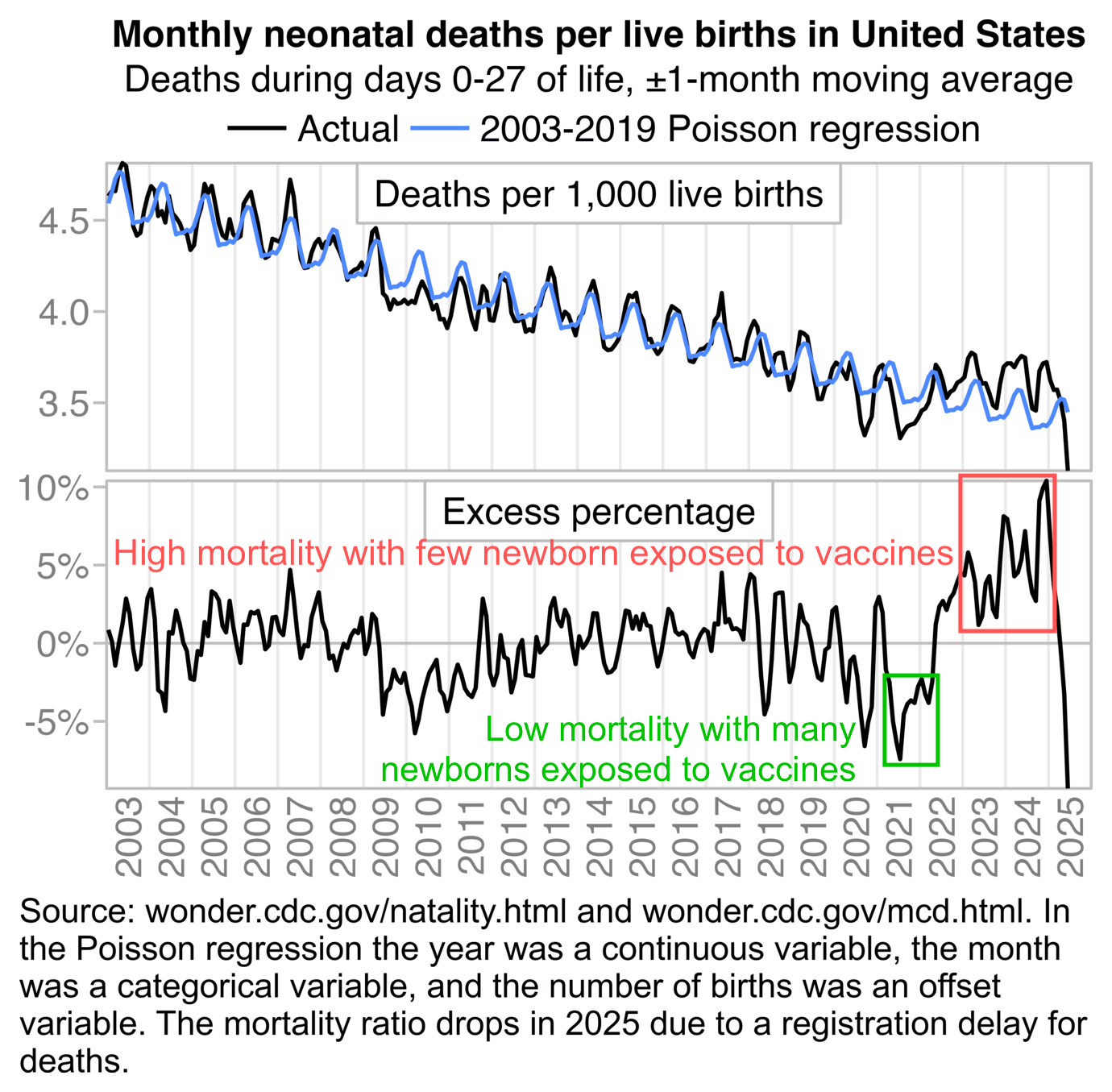
t=fread("https://sars2.net/f/wonderinfantagegroup.csv")
t=t[age!="28-364 days",.(dead=sum(dead)),.(year,month)]
born=fread("https://sars2.net/f/wonderinfantsexmonthly.csv")[,.(born=sum(born)),.(year,month)]
t=merge(t,born)
t[,dead:=ma(dead,1)][,born:=ma(born,1)]
t=t[!(year==2025&month>6)]
t$base=predict(glm(dead~year+factor(month),poisson,t[year<2020],offset=log(born)),t,type="response")
p=t[,.(x=as.Date(paste(year,month,16,sep="-")),y=c(dead,base)/born*1e3,z=rep(1:2,each=.N))]
p=rbind(p[,facet:=1],p[,.(y=(y[1]/y[2]-1)*100,z=1,facet=2),x])
p[,z:=factor(z,,c("Actual","2003-2019 Poisson regression"))]
p[,facet:=factor(facet,,c("Deaths per 1,000 live births","Excess percentage"))]
xstart=as.Date("2003-1-1");xend=as.Date("2026-1-1");xbreak=seq(xstart+182,xend,"year")
ylim=p[,.(min=min(y),max=max(y)),facet]
ggplot(p)+
facet_wrap(~facet,ncol=1,dir="v",scales="free_y")+
geom_vline(xintercept=seq(xstart,xend,"year"),color="gray91",linewidth=.4)+
geom_segment(data=ylim[2],aes(xstart,0),xend=xend,yend=0,color="gray75",linewidth=.4)+
geom_rect(data=ylim,aes(ymin=min,ymax=max),xmin=xstart,xmax=xend,lineend="square",linejoin="mitre",fill=NA,color="gray75",linewidth=.4)+
geom_line(aes(x,y,color=z),linewidth=.6)+
geom_label(data=ylim,aes(mean(c(xstart,xend)),max,label=facet),vjust=1,label.r=unit(0,"pt"),label.padding=unit(5,"pt"),label.size=.4,color="gray75",size=3.87)+
geom_label(data=ylim,aes(mean(c(xstart,xend)),max,label=facet),vjust=1,label.r=unit(0,"pt"),label.padding=unit(5,"pt"),label.size=0,fill=NA,size=3.87)+
labs(x=NULL,y=NULL,title="Monthly neonatal deaths per live births in United States",subtitle="Deaths during days 0-27 of life, ±1-month moving average")+
scale_x_continuous(limits=c(xstart,xend),breaks=xbreak,labels=year(xbreak))+
scale_y_continuous(breaks=pretty,labels=\(x)if(min(x,na.rm=T)<0)paste0(x,"%")else sprintf("%.1f",x))+
scale_color_manual(values=c("black",hsv(22/36,.7,1)))+
scale_alpha_manual(values=c(1,.4))+
coord_cartesian(clip="off",expand=F)+
theme(axis.text=element_text(size=11,color="gray50"),
axis.text.x=element_text(angle=90,vjust=.5,hjust=1),
axis.text.y=element_text(margin=margin(,1)),
axis.ticks=element_line(linewidth=.4,color="gray75"),
axis.ticks.length.x=unit(0,"pt"),
axis.ticks.length.y=unit(4,"pt"),
legend.background=element_blank(),
legend.box.spacing=unit(0,"pt"),
legend.direction="horizontal",
legend.key=element_blank(),
legend.key.height=unit(11,"pt"),
legend.key.width=unit(22,"pt"),
legend.margin=margin(,,5),
legend.position="top",
legend.spacing.x=unit(1,"pt"),
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=11,vjust=.5),
legend.title=element_blank(),
panel.background=element_blank(),
panel.spacing=unit(3,"pt"),
plot.subtitle=element_text(hjust=.5,margin=margin(,,3)),
plot.title=element_text(size=11,hjust=.5,face=2,margin=margin(,,3)),
strip.background=element_blank(),
strip.text=element_blank())
ggsave("1.png",width=4.6,height=3.7,dpi=300*4)
In 2018-2025, CDC WONDER returned a total of 158,615 deaths at age 0-4 with the underlying cause A-R or 999--999, but 99,194 of the deaths were neonatal deaths, so about 63% of the deaths in Ethical Skeptic's plot are neonatal deaths.
In England and Wales, the neonatal mortality rate was also clearly reduced in 2020: [https://x.com/UncleJo46902375/status/1962565388446695774, https://www.ons.gov.uk/peoplepopulationandcommunity/birthsdeathsandmarriages/deaths/datasets/infantmortalitybirthcohorttablesinenglandandwales]
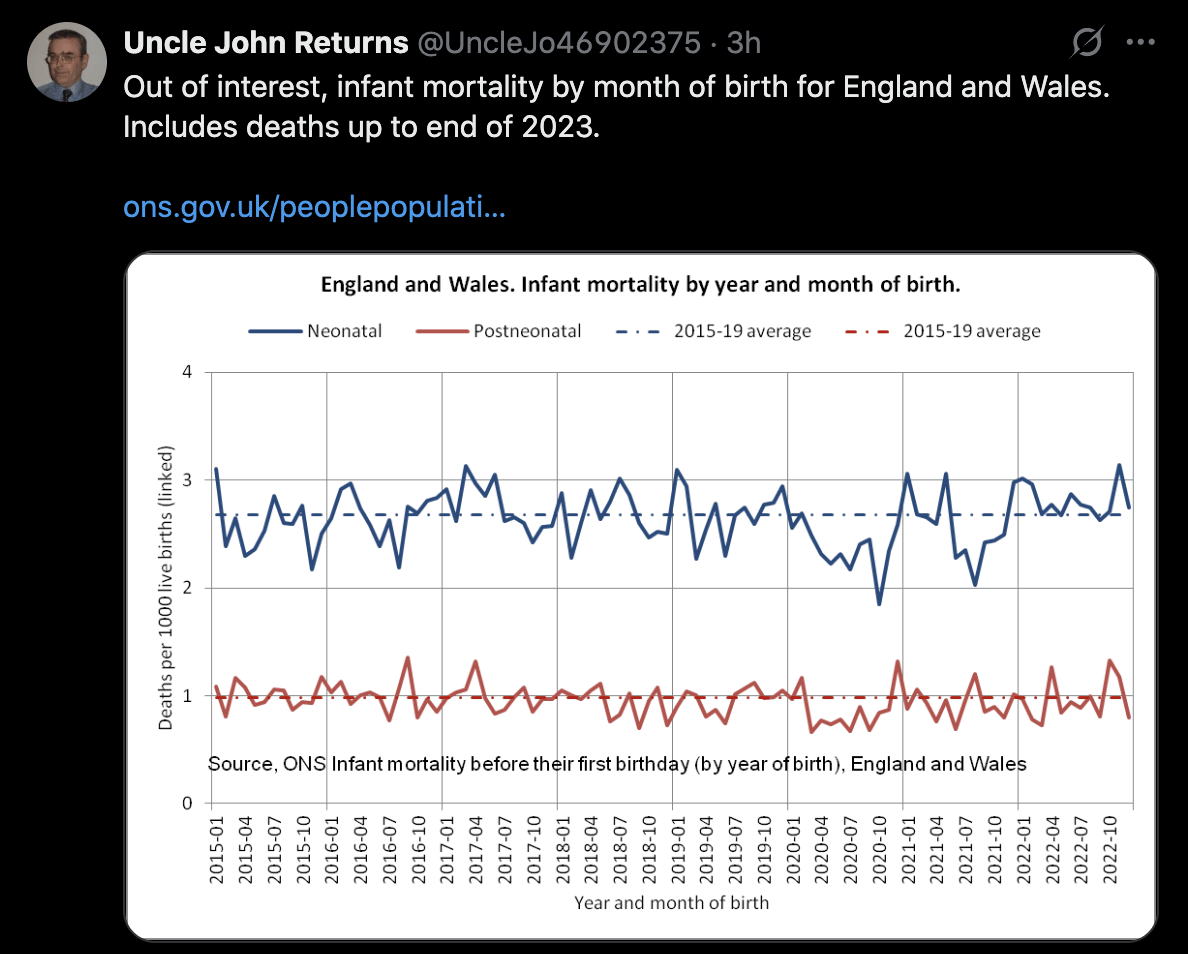
Clare Craig posted this thread (where she looked at deaths from all causes, so she didn't exclude external causes or COVID): [https://x.com/ClareCraigPath/status/1958564816877916348]
I can sort of replicate some of this based on CDC data.
@ethicalskeptic has factored in a pull forward effect for later part of graph.
This is the idea that there should be fewer deaths after a period of excess dying.
It is not unreasonable to include that.
Here is the raw data and trendline.
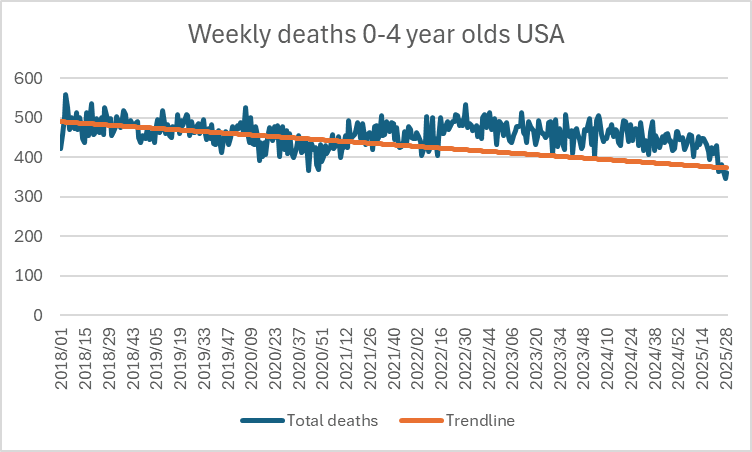
Using a baseline of the trend for all of 2018-2020 gets closer to his/hers.
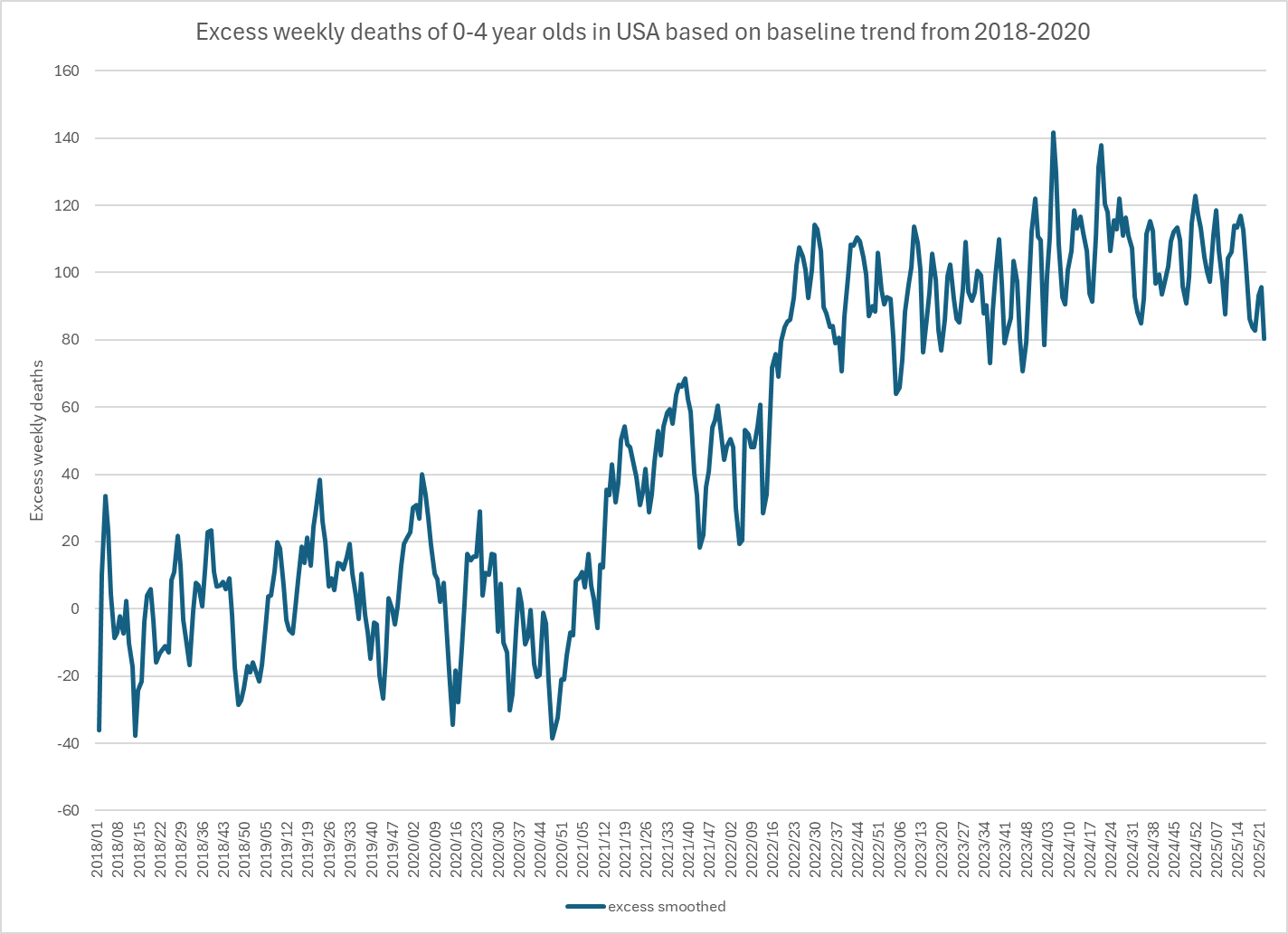
I pointed out to her that there was an unusually low number of deaths in 2020, and that when I reverse engineered Ethical Skeptic's baseline, the slope of his baseline matched the actual deaths in 2018-2019, but it took a sudden dive downwards between 2019 and 2020, so ES seems to have rather fitted his baseline against 2018-2019 and not 2018-2020, but he applied some additional downwards adjustment to the baseline starting from 2020.
I also pointed out that in 2020-2024 in the United States, about 84% of all deaths in ages 0-4 were at age 0, so the plot by ES mostly shows infant mortality, and therefore it's not reasonable to apply extreme PFE adjustment to the baseline.
ES posted these replies to the thread, where he indicated he did not apply PFE adjustment, and he showed that he excluded deaths from external causes and COVID unlike Craig: [https://x.com/EthicalSkeptic/status/1958572434690338818]
Clare Craig also got negative excess deaths for ages 1-4: [https://x.com/ClareCraigPath/status/1958601433055391815]
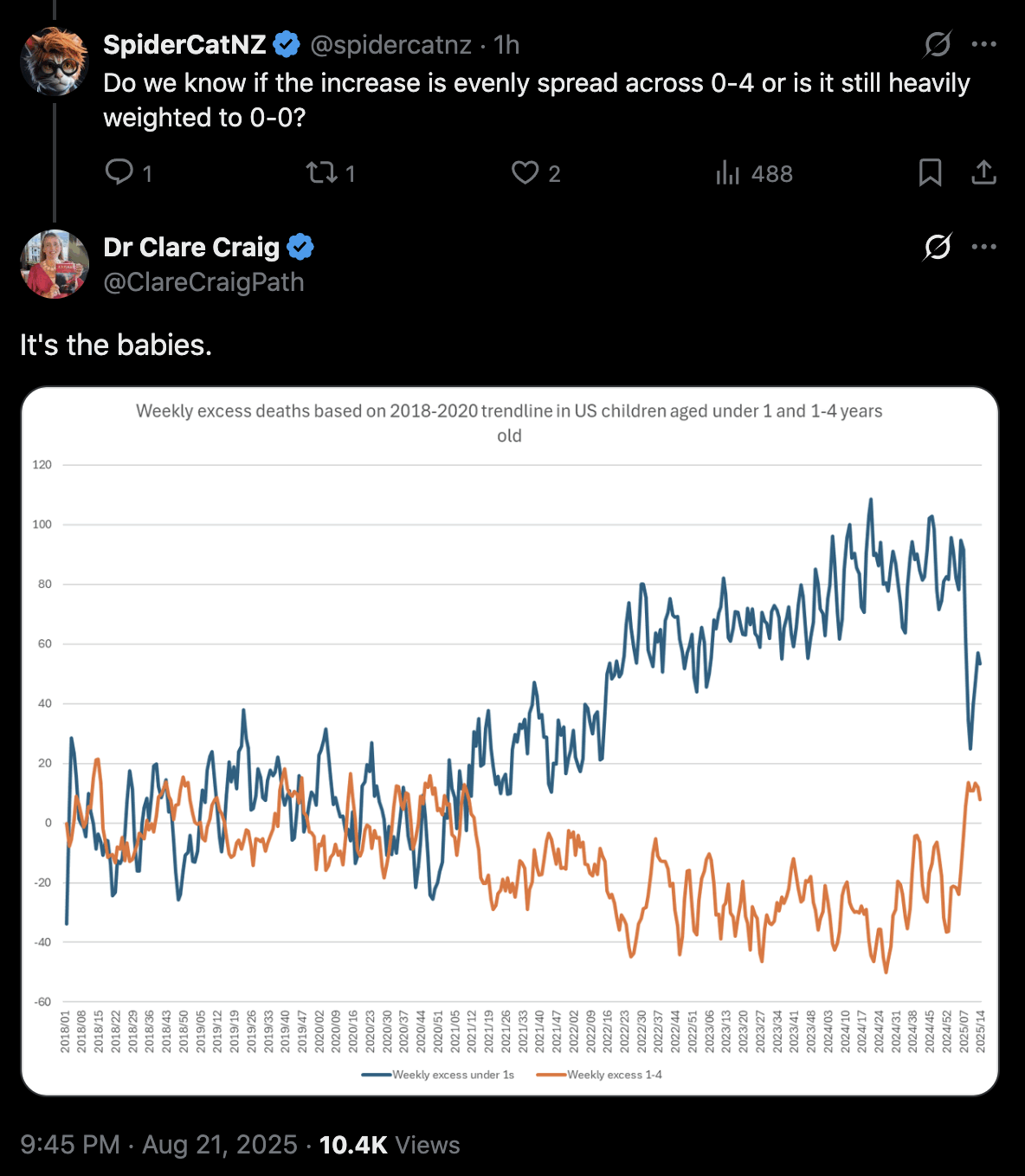
Some people claimed that Clare Craig was able to reproduce Ethical Skeptic's results, but ES claimed that he used a 2018-2019 baseline in his plot where he got 77% excess deaths in 2025, and in Clare Craig's plot where she used a 2018-2019 baseline, towards the end of her x-axis she got an an average of about 430 deaths per week against a baseline of about 380 deaths, so she only got about 13% excess deaths:
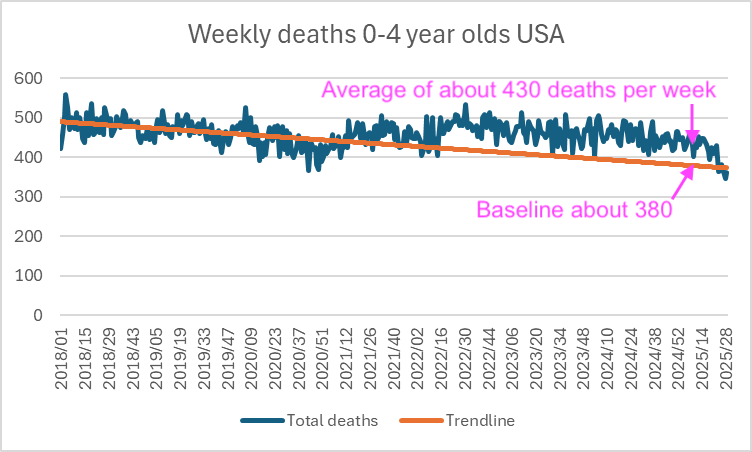
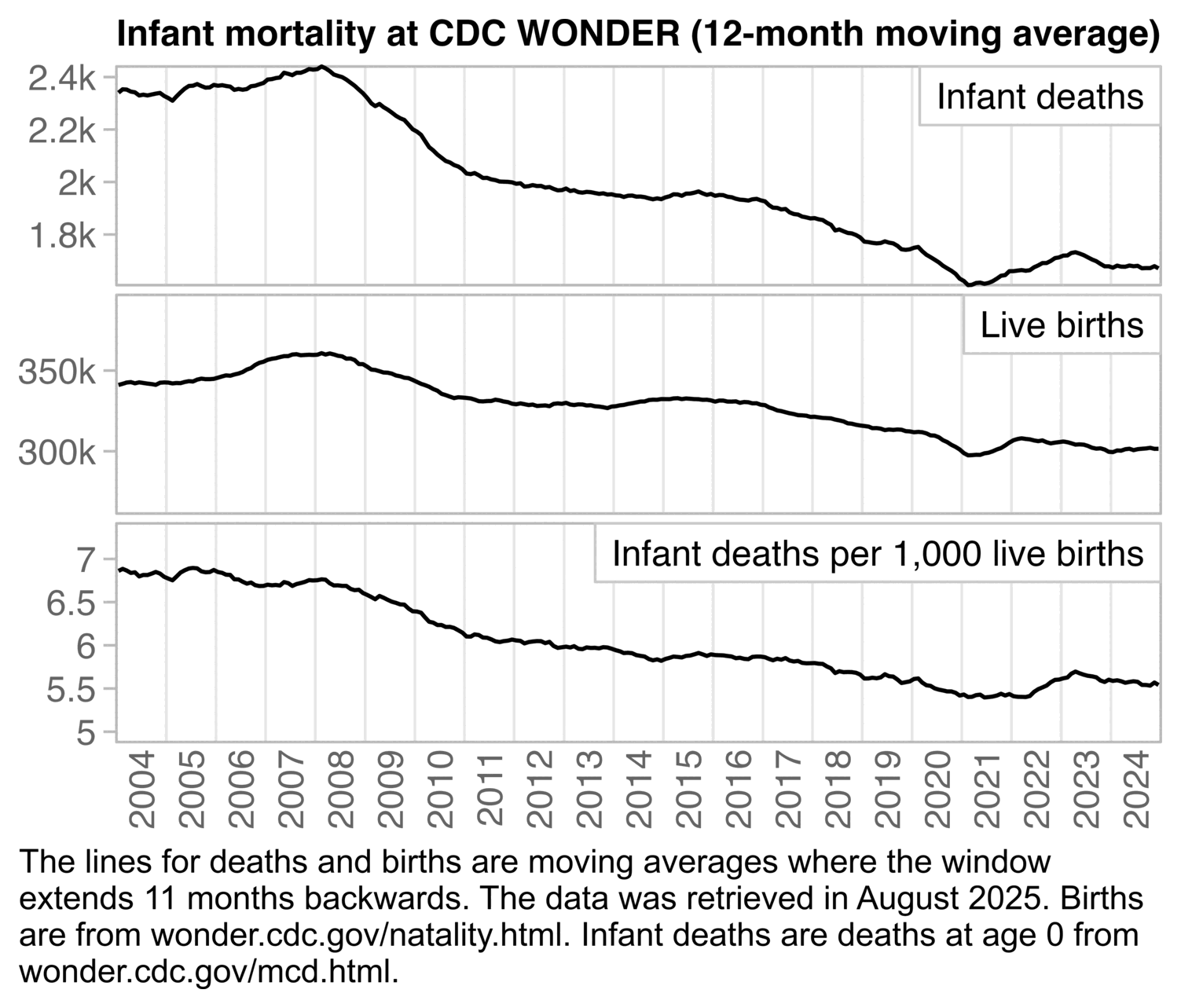
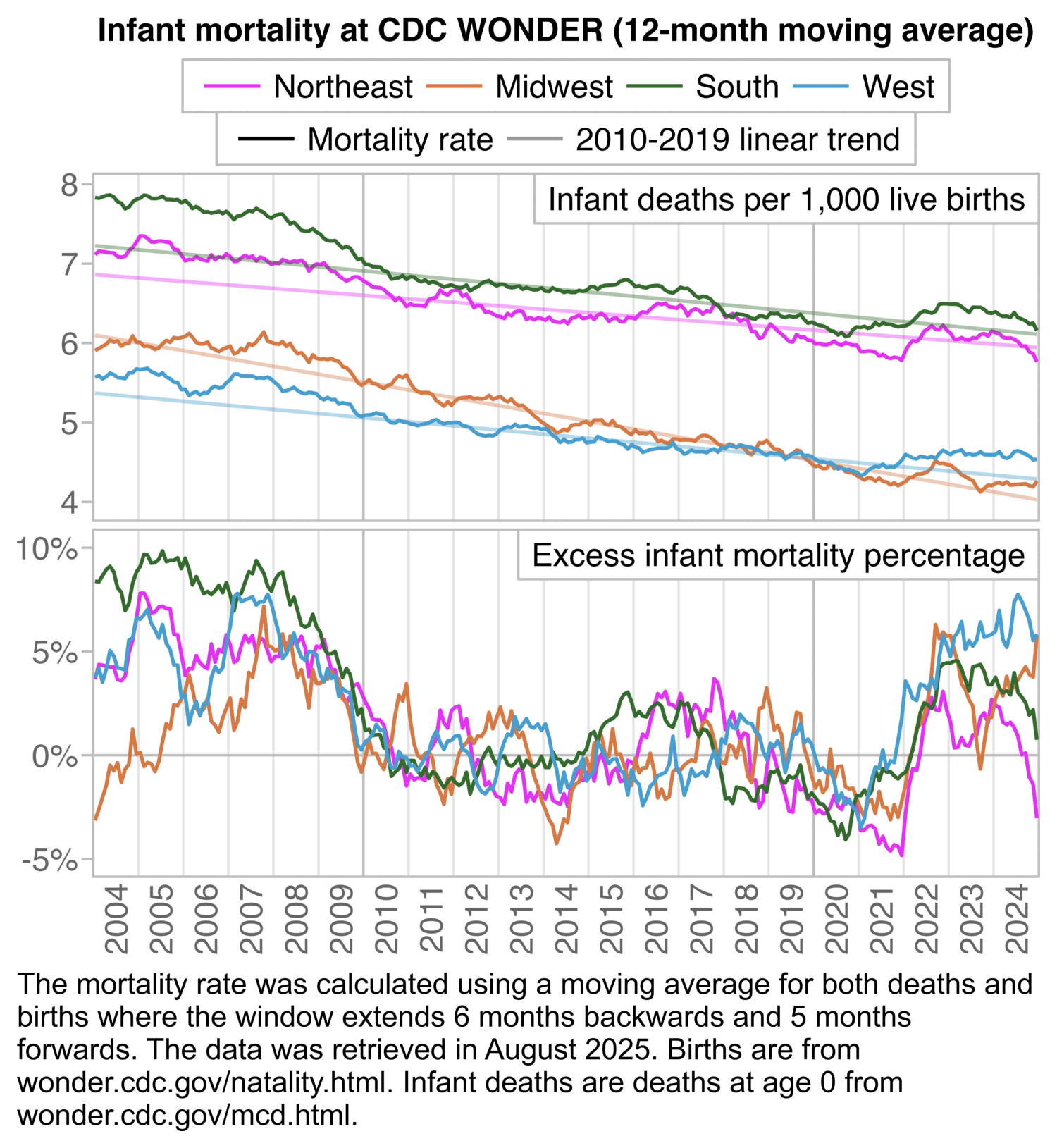
In the next plot that shows a 12-month backwards moving average of monthly data, the deaths per live births starts to increase in mid-2021 and it peaks in mid-2023. The peak might feasibly be due to COVID or vaccines or both:
ma=\(x,b=1,f=b){x[]=rowMeans(embed(c(rep(NA,b),x,rep(NA,f)),f+b+1),na.rm=T);x}
t=fread("https://sars2.net/f/wondernatality.csv")
d=fread("https://sars2.net/f/uspopdeadmonthly.csv")[age==0,.(year,month,dead)]
t=merge(t,d)[,x:=as.Date(paste0(year,"-",month,"-16"))]
t[,born:=ma(born,11,0)][,dead:=ma(dead,11,0)]
p=t[year%in%2004:2024,.(x,y=c(dead,born,dead/born*1e3),z=rep(1:3,each=.N))]
p[,z:=factor(z,,c("Infant deaths","Live births","Infant deaths per 1,000 live births"))]
xstart=as.Date("2004-1-1");xend=as.Date("2025-1-1");xbreak=seq(xstart+182,xend,"year")
ylim=p[,.(min=min(y),max=max(y)),z]
rat=ylim[,max(max/min)]
ylim=ylim[,{x=(rat*min-max)/(1+rat);.(min=min-x,max=max+x,z)}]
ggplot(p)+
facet_wrap(~z,ncol=1,dir="v",scales="free_y")+
geom_vline(xintercept=seq(xstart,xend,"year"),color="gray90",linewidth=.4)+
geom_rect(data=ylim,aes(ymin=min,ymax=max),xmin=xstart,xmax=xend,lineend="square",linejoin="mitre",fill=NA,color="gray75",linewidth=.4)+
geom_line(aes(x,y),linewidth=.6)+
geom_label(data=ylim,aes(xend,max,label=z),vjust=1,hjust=1,label.r=unit(0,"pt"),label.padding=unit(5,"pt"),label.size=.4,size=3.87,fill="white",color="gray80")+
geom_label(data=ylim,aes(xend,max,label=z),vjust=1,hjust=1,label.r=unit(0,"pt"),label.padding=unit(5,"pt"),label.size=0,size=3.87,fill=NA)+
labs(title="Infant mortality at CDC WONDER (12-month moving average)",x=NULL,y=NULL)+
scale_x_continuous(limits=c(xstart,xend),breaks=xbreak,labels=year(xbreak))+
scale_y_continuous(breaks=\(x)pretty(x,4),labels=\(x)ifelse(x>=1e3,paste0(x/1e3,"k"),x))+
coord_cartesian(clip="off",expand=F)+
theme(axis.text=element_text(size=11,color="gray40",margin=margin(2,2,2,2)),
axis.text.x=element_text(angle=90,vjust=.5,hjust=1),
axis.ticks=element_line(linewidth=.4,color="gray75"),
axis.ticks.length.x=unit(0,"pt"),
axis.ticks.length.y=unit(4,"pt"),
panel.background=element_blank(),
panel.grid=element_blank(),
panel.spacing=unit(3,"pt"),
plot.title=element_text(size=11,hjust=.5,face="bold",margin=margin(1,,4)),
strip.background=element_blank(),
strip.text=element_blank())
ggsave("1.png",width=5,height=3.6,dpi=300*4)
Younger age groups had a high number of COVID deaths in 2022 and 2021 relative to 2020, so women aged 15-44 had almost as many UCD COVID deaths in 2022 as 2020:
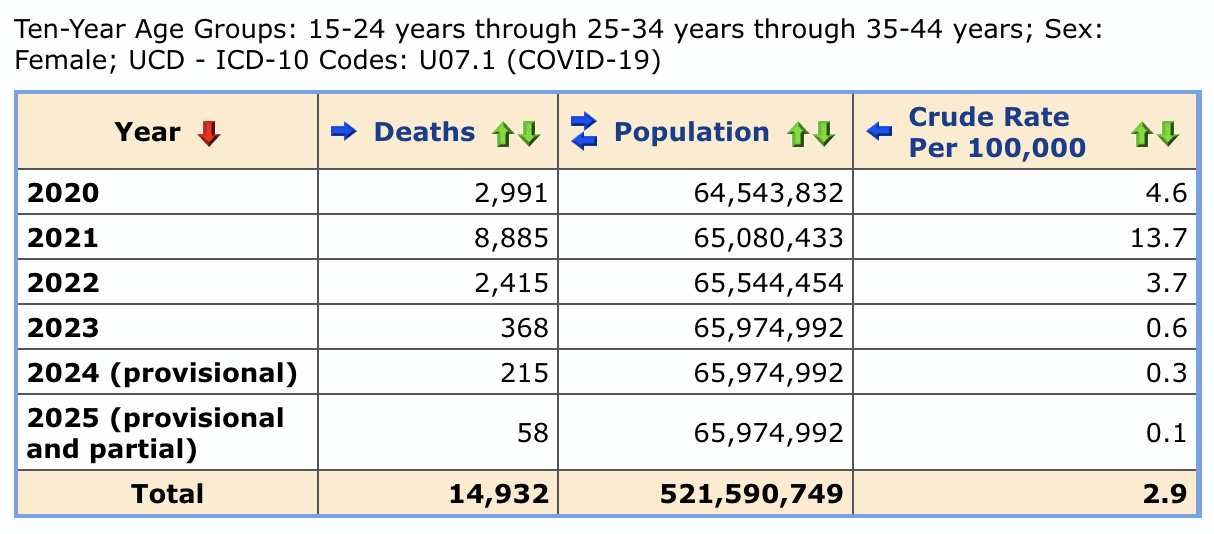
Jean Fisch posted this thread: [https://x.com/Jean__Fisch/status/1958613062971531527]
There is a theory circulating that the 0-4 in the US saw an increase of deaths as of 2021 and "it's the kids of the 1st vaccinated mothers"
If you look at trends, the change happened at the same time for the <1 and the 1-4 yrs old ... who were born in 2017-2020
Not only that ..
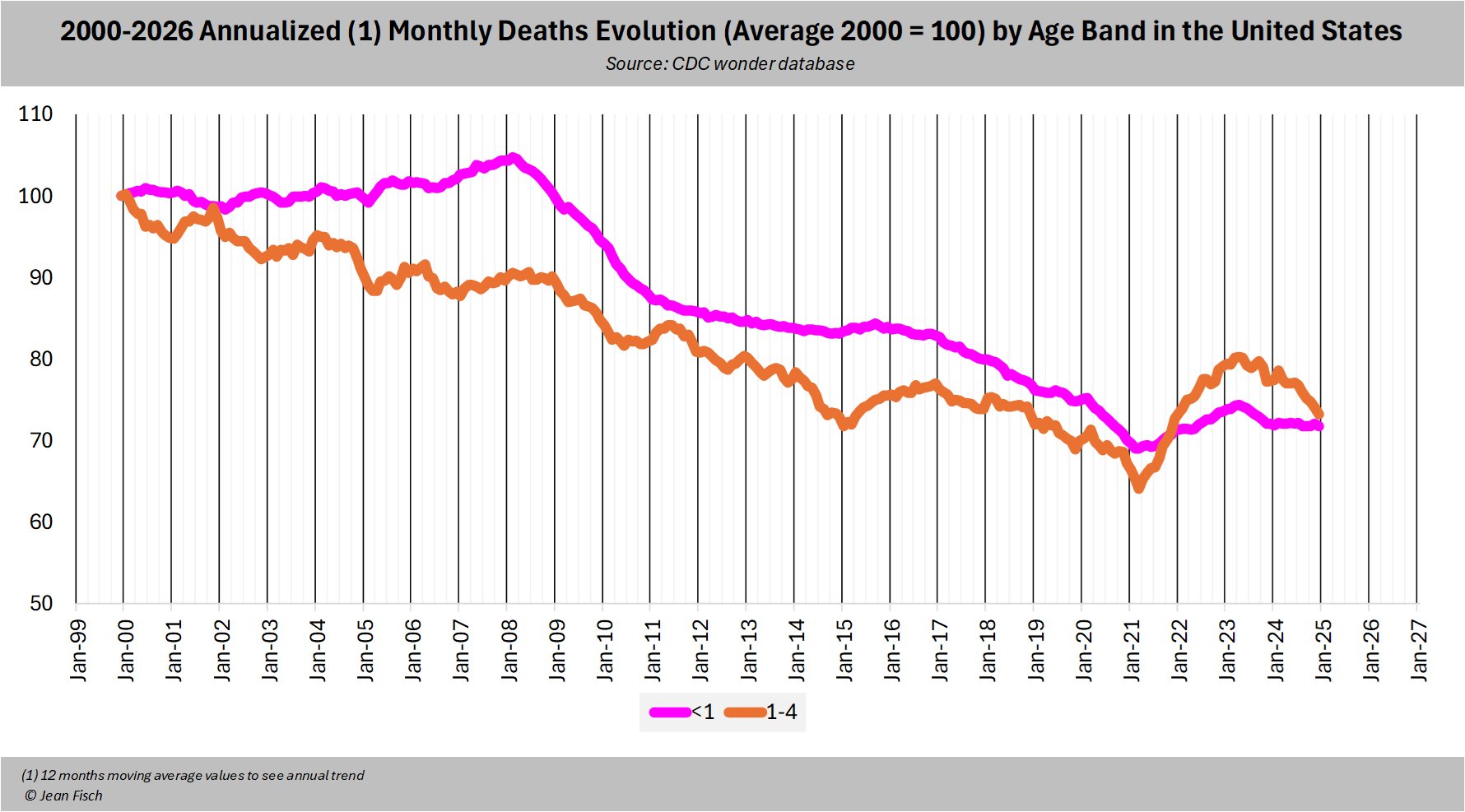
If you could find a way to explain the shift in the 1-4 year old and zoom in on the <1 yr old, you would then have to explain why the shift as of 2021 is most significant in the Census region south, well-known for having the highest vax uptake in the US (not) 😀
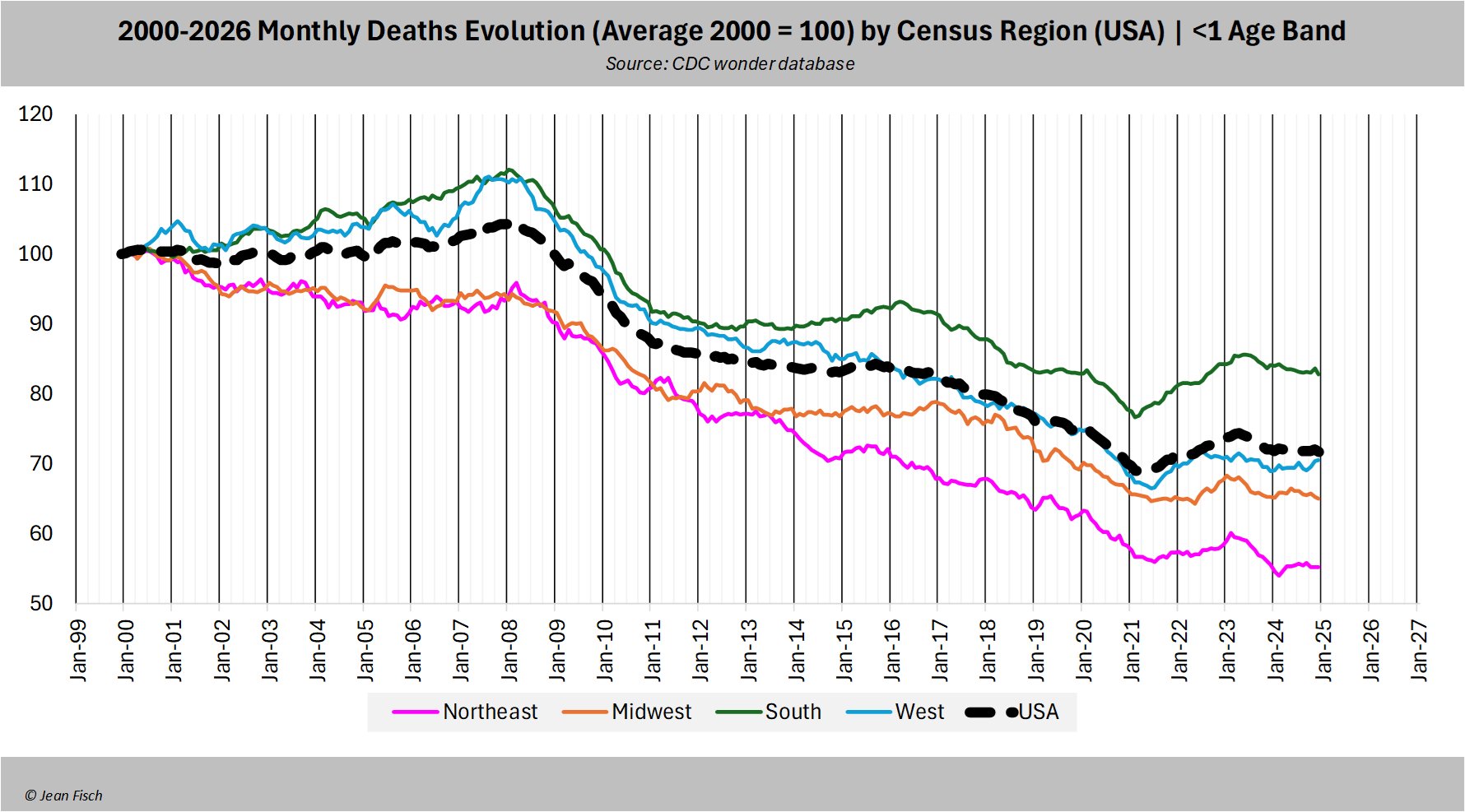
When I looked at deaths per live births relative to a 2010-2019 linear trend, I got the lowest excess infant mortality in the northeastern census region, which probably has the highest vaccination rate:
ma=\(x,b=1,f=b){x[]=rowMeans(embed(c(rep(NA,b),x,rep(NA,f)),f+b+1),na.rm=T);x}
t=fread("https://sars2.net/f/wondernatalityregion.csv")[order(region,year,month)]
t=t[,.(born=ma(born,6,5),dead=ma(dead,6,5),year,month),region]
t[,x:=as.Date(paste0(year,"-",month,"-16"))]
p=t[year%in%2004:2024,.(x,y=dead/born*1e3,z=1,region)]
p=rbind(p,p[,.(x,y=predict(lm(y~x,.SD[year(x)%in%2010:2019]),.SD),z=2),region])
p=rbind(p[,facet:=1],p[,.(y=(y[1]/y[2]-1)*100,z=1,facet=2),.(x,region)])
p[,z:=factor(z,,c("Mortality rate","2010-2019 linear trend"))]
p[,facet:=factor(facet,,c("Infant deaths per 1,000 live births","Excess infant mortality percentage"))]
p[,region:=factor(region,,c("Northeast","Midwest","South","West"))]
xstart=as.Date("2004-1-1");xend=as.Date("2025-1-1");xbreak=seq(xstart+182,xend,"year")
ylim=p[,{x=extendrange(y,,.07);.(min=x[1],max=x[2])},facet]
ggplot(p)+
facet_wrap(~facet,ncol=1,dir="v",scales="free_y")+
geom_vline(xintercept=seq(xstart,xend,"year"),color="gray90",linewidth=.4)+
geom_vline(xintercept=as.Date(paste0(c(2010,2020),"-1-1")),color="gray75",linewidth=.4)+
geom_segment(data=ylim[2],x=xstart,xend=xend,y=0,yend=0,color="gray75",linewidth=.4)+
geom_rect(data=ylim,aes(ymin=min,ymax=max),xmin=xstart,xmax=xend,lineend="square",linejoin="mitre",fill=NA,color="gray75",linewidth=.4)+
geom_line(aes(x,y,color=region,alpha=z),linewidth=.6)+
geom_label(data=ylim,aes(xend,max,label=facet),hjust=1,vjust=1,label.r=unit(0,"pt"),label.padding=unit(4.5,"pt"),label.size=.4,size=3.87,fill="white",color="gray75")+
geom_label(data=ylim,aes(xend,max,label=facet),hjust=1,vjust=1,label.r=unit(0,"pt"),label.padding=unit(4.5,"pt"),label.size=0,size=3.87,fill=NA)+
labs(title="Infant mortality at CDC WONDER (12-month moving average)",x=NULL,y=NULL)+
scale_x_continuous(limits=c(xstart,xend),breaks=xbreak,labels=year(xbreak))+
scale_y_continuous(breaks=\(x)pretty(x,4),labels=\(x)if(min(x,na.rm=T)<0)paste0(x,"%")else x)+
scale_color_manual(values=c(hcl(225,100,50),hcl(55,70,50),"black",hcl(135,100,50)))+
scale_alpha_manual(values=c(1,.4))+
coord_cartesian(clip="off",expand=F)+
theme(axis.text=element_text(size=11,color="gray40"),
axis.text.x=element_text(angle=90,vjust=.5,hjust=1),
axis.text.y=element_text(margin=margin(,1)),
axis.ticks=element_line(linewidth=.4,color="gray75"),
axis.ticks.length.x=unit(0,"pt"),
axis.ticks.length.y=unit(4,"pt"),
legend.background=element_rect(color="gray75",linewidth=.4),
legend.box="vertical",
legend.box.margin=margin(,,3),
legend.box.spacing=unit(0,"pt"),
legend.direction="horizontal",
legend.key=element_blank(),
legend.key.height=unit(12,"pt"),
legend.key.width=unit(24,"pt"),
legend.margin=margin(3,5,3,3),
legend.position="top",
legend.spacing.x=unit(2,"pt"),
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=11),
legend.title=element_blank(),
panel.background=element_blank(),
panel.grid=element_blank(),
panel.spacing=unit(3,"pt"),
plot.title=element_text(size=11,hjust=.5,face="bold",margin=margin(1,,4)),
strip.background=element_blank(),
strip.text=element_blank())
ggsave("1.png",width=5,height=4.6,dpi=300*4)
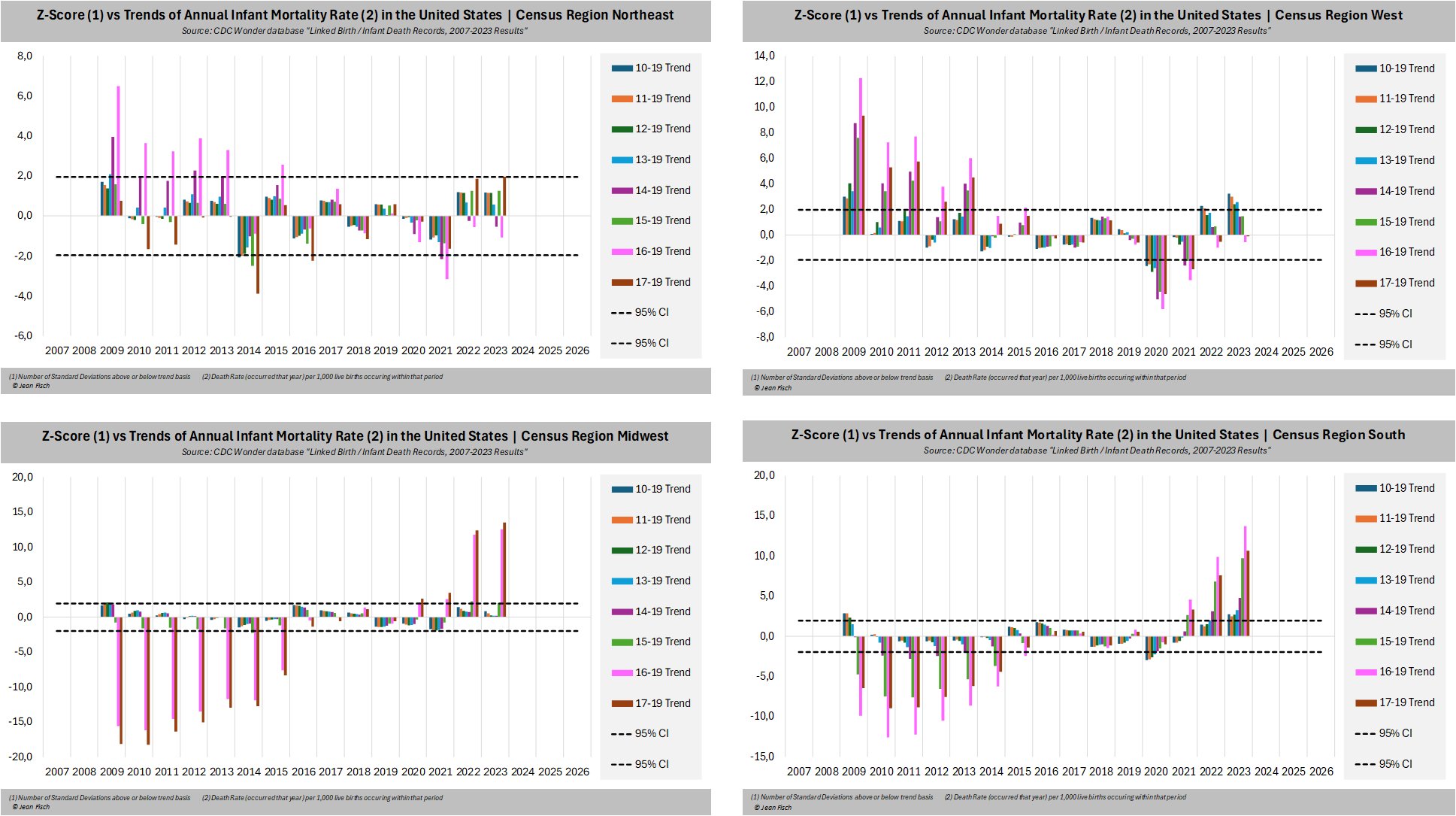
The next plot shows that even with 8 different baselines to choose from, the northeastern census region never got statistically significant excess infant mortality in 2022 or 2023: [https://x.com/Jean__Fisch/status/1964326908071448814]
In EU the excess infant mortality rate relative to a 2012-2019 trend was about -2% in 2021, 3% in 2022, and 4% in 2023. But many of the countries with the highest excess infant mortality rate in 2021-2023 were Eastern European countries with a low vaccination rate:
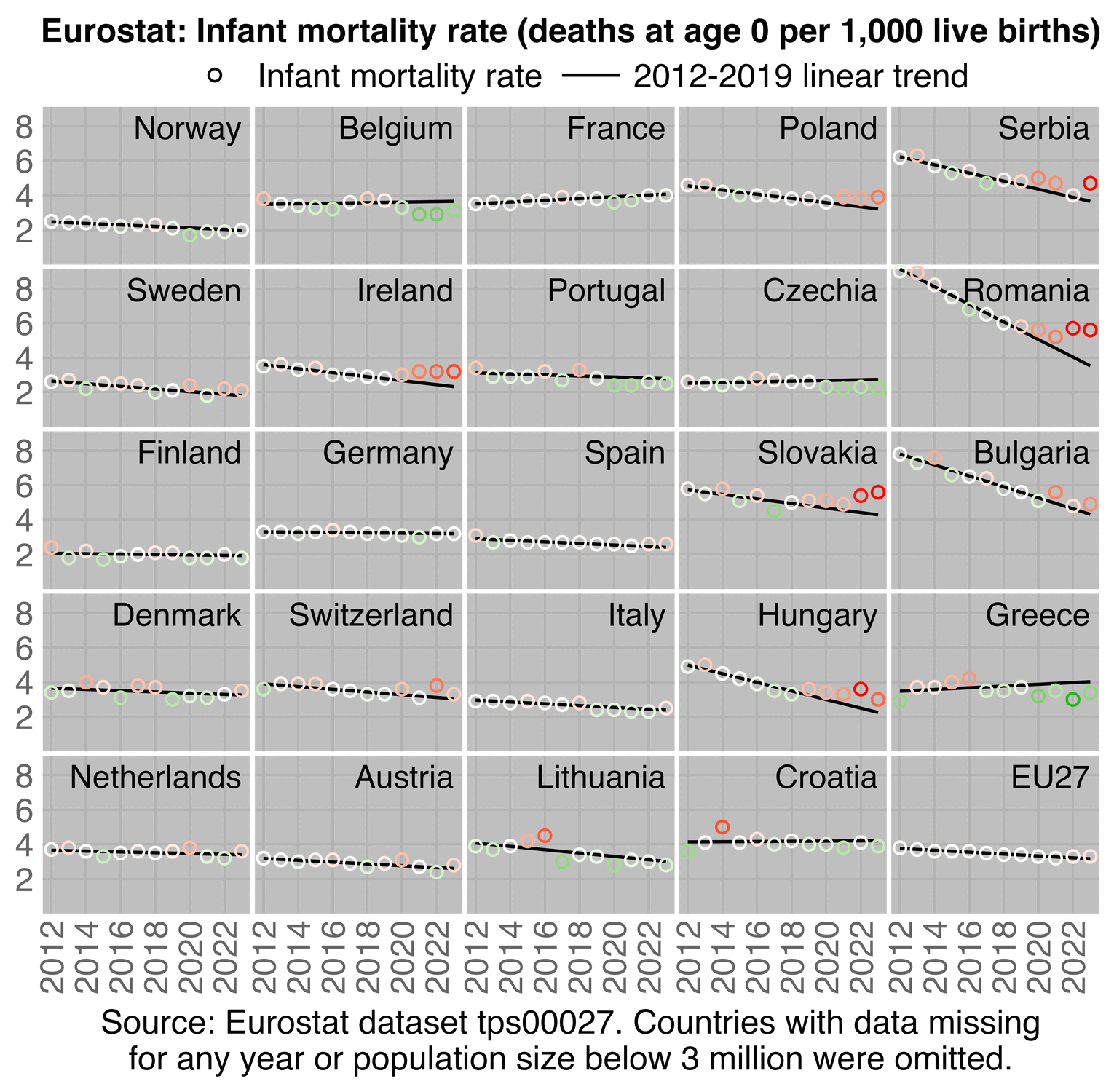
download.file("https://ec.europa.eu/eurostat/api/dissemination/sdmx/2.1/data/tps00027?format=TSV","tps00027.tsv")
country=fread("code,name
NO,Norway
SE,Sweden
FI,Finland
DK,Denmark
NL,Netherlands
BE,Belgium
IE,Ireland
DE,Germany
CH,Switzerland
AT,Austria
FR,France
PT,Portugal
ES,Spain
IT,Italy
LT,Lithuania
PL,Poland
CZ,Czechia
SK,Slovakia
HU,Hungary
HR,Croatia
RS,Serbia
RO,Romania
BG,Bulgaria
EL,Greece
EU27_2020,EU27")
t=fread("tps00027.tsv",na=":",head=T)
t=t[,.(z=sub(".*,","",t[[1]]),y=unlist(.SD[,-1]),x=rep(2012:2023,each=.N))]
t[,y:=as.double(sub(" .*","",y))]
t=na.omit(t)
p=t[z%in%t[,.N,z][N==max(N),z]]
p=p[z%in%country$code]
p[,z:=factor(z,country$code,country$name)]
p=p[,.(x,y,base=predict(lm(y~x,.SD[x%in%2012:2019]),.SD)),z]
xstart=2012;xend=2023;xbreak=seq(xstart,xend,2)
yend=p[,max(y,base,na.rm=T)];ybreak=1:5*2
ggplot(p)+
facet_wrap(~z,ncol=5,dir="v")+
geom_line(aes(x,base,linetype="2012-2019 linear trend"),linewidth=.5)+
geom_point(aes(x,y,color=pmax(pmin(y-base,1),-1),shape="Infant mortality rate"),stroke=.6,size=1.5)+
geom_text(data=p[rowid(z)==1],aes(label=z),vjust=1.4,hjust=1,color="black",x=xend,y=yend,size=3.7)+
labs(title="Eurostat: Infant mortality rate (deaths at age 0 per 1,000 live births)",caption="Source: Eurostat dataset tps00027. Countries with data missing\nfor any year or population size below 3 million were omitted.",x=NULL,y=NULL)+
scale_x_continuous(limits=c(xstart-.5,xend+.5),breaks=xbreak)+
scale_y_continuous(breaks=ybreak)+
scale_color_gradientn(colors=c("red","white","#00bb00"),breaks=c(-1,0,1),limits=c(-1,1),guide="none")+
scale_linetype_manual(values=1)+
scale_shape_manual(values=1)+
coord_cartesian(ylim=c(0,yend),clip="off",expand=F)+
guides(shape=guide_legend(order=1),keywidth=unit(10,"pt"))+
theme(axis.text=element_text(size=11,color="gray40",margin=margin(2,2,2,2)),
axis.text.x=element_text(angle=90,vjust=.5,hjust=1),
axis.text.y=element_text(margin=margin(,3)),
axis.ticks=element_line(linewidth=.4),
axis.ticks.length=unit(0,"pt"),
axis.title=element_text(size=11,face=2),
legend.background=element_blank(),
legend.box.spacing=unit(0,"pt"),
legend.direction="horizontal",
legend.justification="center",
legend.key=element_blank(),
legend.key.height=unit(13,"pt"),
legend.key.width=unit(24,"pt"),
legend.margin=margin(,,4),
legend.position="top",
legend.spacing.x=unit(2,"pt"),
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=11,vjust=.5),
legend.title=element_blank(),
panel.background=element_rect(fill="gray75"),
panel.grid=element_blank(),
panel.grid.major=element_line(linewidth=.4,color="gray70"),
panel.spacing=unit(1.5,"pt"),
plot.margin=margin(5,5,5,5),
plot.caption=element_text(size=11,hjust=.5,margin=margin(5),lineheight=.9),
plot.title=element_text(size=11,hjust=.5,face="bold",margin=margin(1,,2)),
strip.background=element_blank(),
strip.text=element_blank())
ggsave("1.png",width=5.1,height=5,dpi=300*4)
In many developing countries the decline in infant mortality rate has become less steep over time, and in some countries the infant mortality rate had already flattened out by the 2010s. So relative to the long-term curved trend, a 2010-2019 linear baseline might also be too steep in the United States by the 2020s: [https://platform.who.int/data/maternal-newborn-child-adolescent-ageing/indicator-explorer-new/MCA/infant-mortality-rate-%28per-1000-live-births%29]
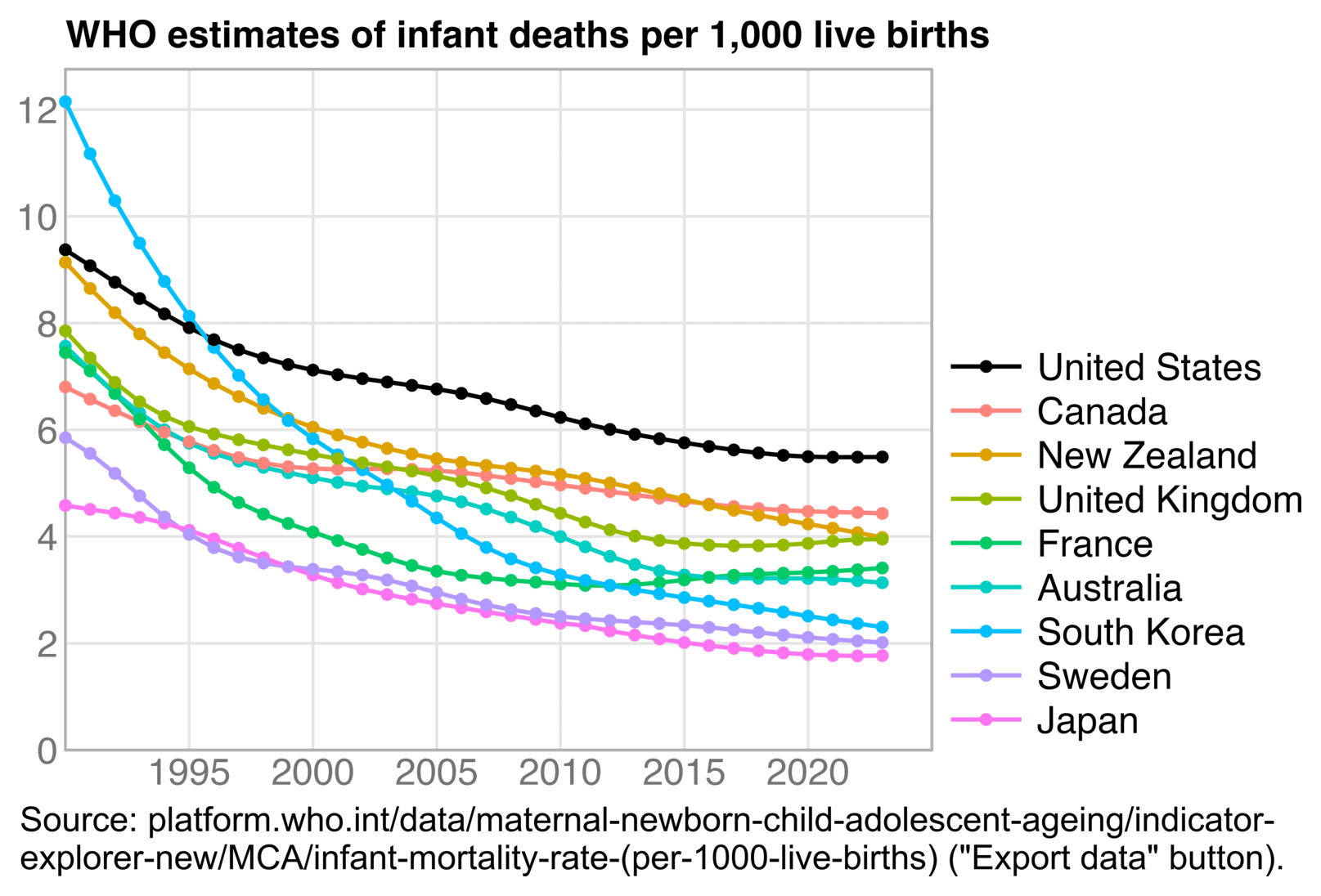
t=setDT(read_excel("infantmortalitywhocomplete.xlsx",sheet=2,guess_max=Inf))
s=t[Country%like%"^Republic of Korea|United States|Sweden|Japan|Australia|Zeala|Canada|United Kingdom|France"&Sex=="Both sexes"]
p=s[,.(z=Country,y=`Value Numeric`,x=as.integer(Year))][x>=1990]
p[z=="United States of America",z:="United States"]
p[z=="Republic of Korea",z:="South Korea"]
p[z%like%"United King",z:="United Kingdom"]
p[,z:=factor(z,p[x==2023][order(-y),z])]
xstart=1990;xend=2025;xbreak=seq(1995,xend-1,5)
ylim=c(0,max(p$y)*1.05);ybreak=pretty(ylim,7)
ggplot(p)+
annotate("rect",xmin=xstart,xmax=xend,ymin=ylim[1],ymax=ylim[2],linewidth=.4,lineend="square",fill=NA,color="gray70")+
geom_line(aes(x,y,color=z),linewidth=.6)+
geom_point(aes(x,y,color=z),size=1.1)+
labs(x=NULL,y=NULL,title="WHO estimates of infant deaths per 1,000 live births")+
scale_x_continuous(limits=c(xstart,xend),breaks=xbreak)+
scale_y_continuous(limits=ylim,breaks=ybreak,labels=kim)+
coord_cartesian(clip="off",expand=F)+
scale_color_manual(values=c("black",rainbow_hcl(nlevels(p$z)-1,100,70,start=15)))+
theme(axis.text=element_text(size=11,color="gray45"),
axis.ticks=element_line(linewidth=.4,color="black"),
axis.ticks.length=unit(0,"pt"),
legend.background=element_blank(),
legend.box.spacing=unit(0,"pt"),
legend.justification=c(0,.03),
legend.key=element_blank(),
legend.key.height=unit(13,"pt"),
legend.key.width=unit(26,"pt"),
legend.margin=margin(,,,3),
legend.position="right",
legend.spacing.x=unit(2,"pt"),
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=11),
legend.title=element_blank(),
panel.background=element_blank(),
panel.grid.major=element_line(linewidth=.4,color="gray90"),
plot.title=element_text(size=11,hjust=0,face=2,margin=margin(,,4)))
ggsave("1.png",width=5.4,height=3.3,dpi=300*4)
In England and Wales the infant mortality rate had already flattened out in the 2010s, so in the US the flattening out in the 2020s might be part of a similar long-term curved trend: [https://www.ons.gov.uk/peoplepopulationandcommunity/birthsdeathsandmarriages/deaths/datasets/childmortalitystatisticschildhoodinfantandperinatalchildhoodinfantandperinatalmortalityinenglandandwales]
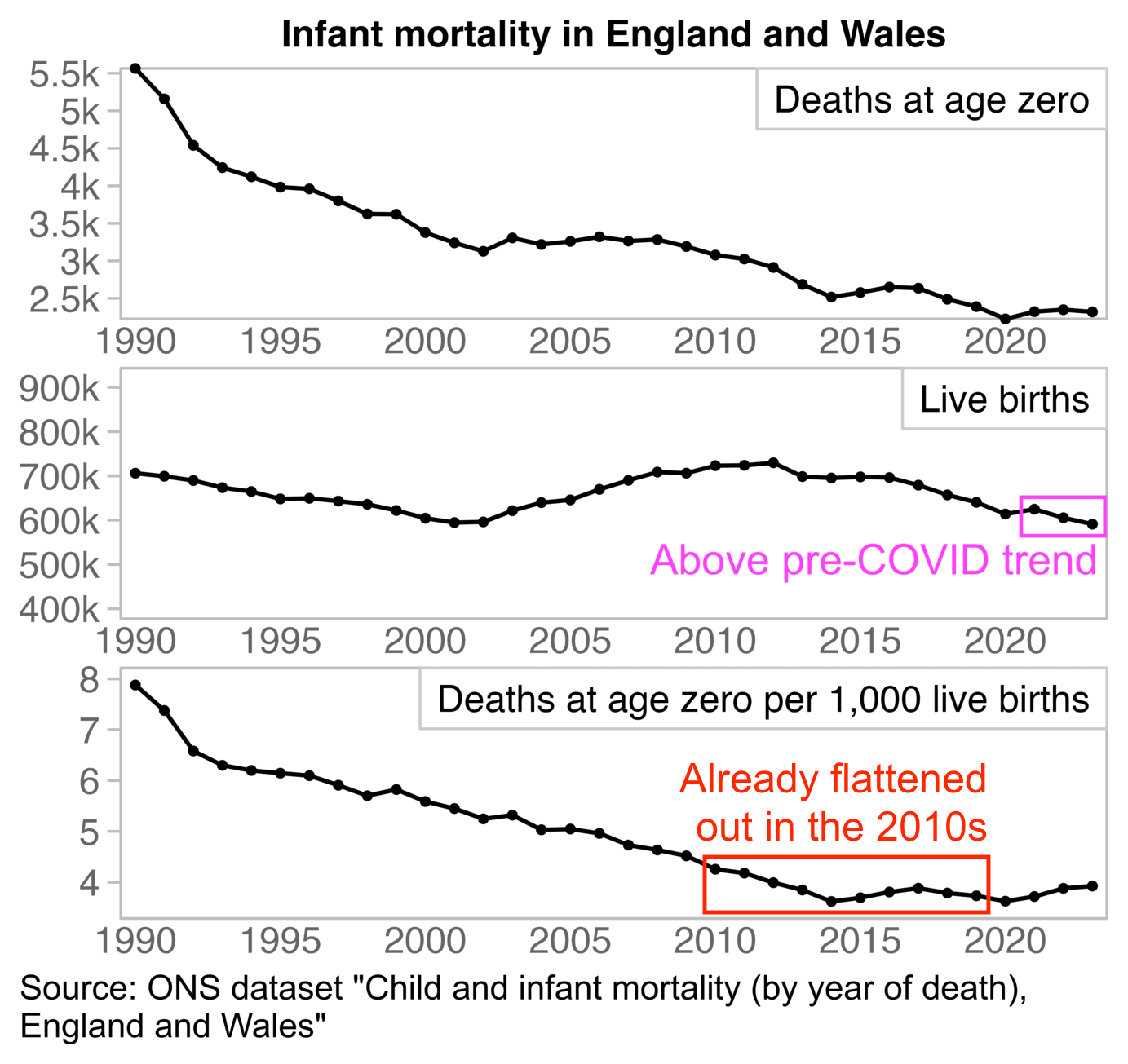
download.file("https://www.ons.gov.uk/file?uri=/peoplepopulationandcommunity/birthsdeathsandmarriages/deaths/datasets/childmortalitystatisticschildhoodinfantandperinatalchildhoodinfantandperinatalmortalityinenglandandwales/2023/cim2023deathcohortworkbook.xlsx","cim2023deathcohortworkbook.xlsx")
t=setDT(read_excel("cim2023deathcohortworkbook.xlsx",sheet=5,skip=9))
p=t[,.(x=Year,born=`Live births`,dead=`Infant under 1 year`)]
p=p[,.(x,y=c(dead,born,dead/born*1e3),z=rep(1:3,each=.N))]
p[,z:=factor(z,,c("Deaths at age zero","Live births","Deaths at age zero per 1,000 live births"))]
xstart=1990;xend=2023;xbreak=seq(xstart,xend,5)
p=p[x%in%xstart:xend]
ylim=p[,.(min=min(y),max=max(y)),z]
rat=ylim[,max(max/min)]
ylim=ylim[,{x=(rat*min-max)/(1+rat);.(min=min-x,max=max+x,z)}]
ggplot(p)+
facet_wrap(~z,ncol=1,dir="v",scales="free")+
geom_rect(data=ylim,aes(ymin=min,ymax=max),xmin=xstart-.5,xmax=xend+.5,lineend="square",linejoin="mitre",fill=NA,color="gray75",linewidth=.4)+
geom_line(aes(x,y),linewidth=.6)+
geom_point(aes(x,y),stroke=0,size=1.5)+
geom_label(data=ylim,aes(xend+.5,max,label=z),vjust=1,hjust=1,label.r=unit(0,"pt"),label.padding=unit(5,"pt"),label.size=.4,size=3.87,fill="white",color="gray80")+
geom_label(data=ylim,aes(xend+.5,max,label=z),vjust=1,hjust=1,label.r=unit(0,"pt"),label.padding=unit(5,"pt"),label.size=0,size=3.87,fill=NA)+
labs(title="Infant mortality in England and Wales",x=NULL,y=NULL)+
scale_x_continuous(limits=c(xstart-.5,xend+.5),breaks=xbreak)+
scale_y_continuous(breaks=pretty,labels=\(x)ifelse(x>=1e3,paste0(x/1e3,"k"),x))+
scale_alpha_manual(values=c(1,0))+
coord_cartesian(clip="off",expand=F)+
theme(axis.text=element_text(size=11,color="gray40",margin=margin(2,2,2,2)),
axis.ticks=element_line(linewidth=.4,color="gray75"),
axis.ticks.length.x=unit(0,"pt"),
axis.ticks.length.y=unit(4,"pt"),
panel.background=element_blank(),
panel.grid=element_blank(),
panel.spacing=unit(2,"pt"),
plot.subtitle=element_text(size=11,hjust=.5,margin=margin(,,4)),
plot.title=element_text(size=11,hjust=.5,face="bold",margin=margin(,,4)),
strip.background=element_blank(),
strip.text=element_blank())
ggsave("1.png",width=4.6,height=4,dpi=300*4)
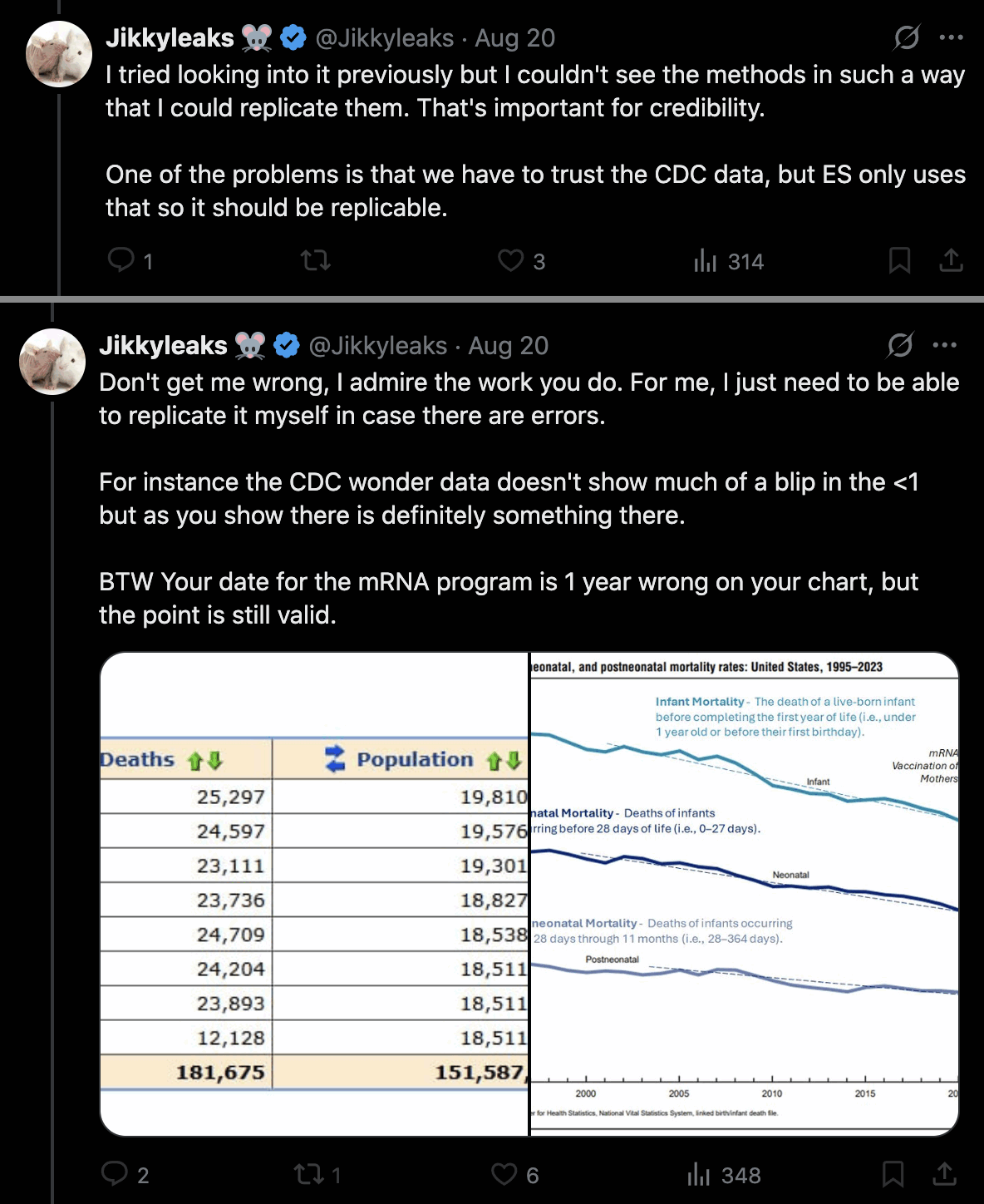
Jikkyleaks also said that Ethical Skeptic should document his methodology. He got about 6% lower deaths in ages 0-4 in 2024 than 2018, even though he looked at deaths from all causes: [https://x.com/Jikkyleaks/status/1958016656573362408]
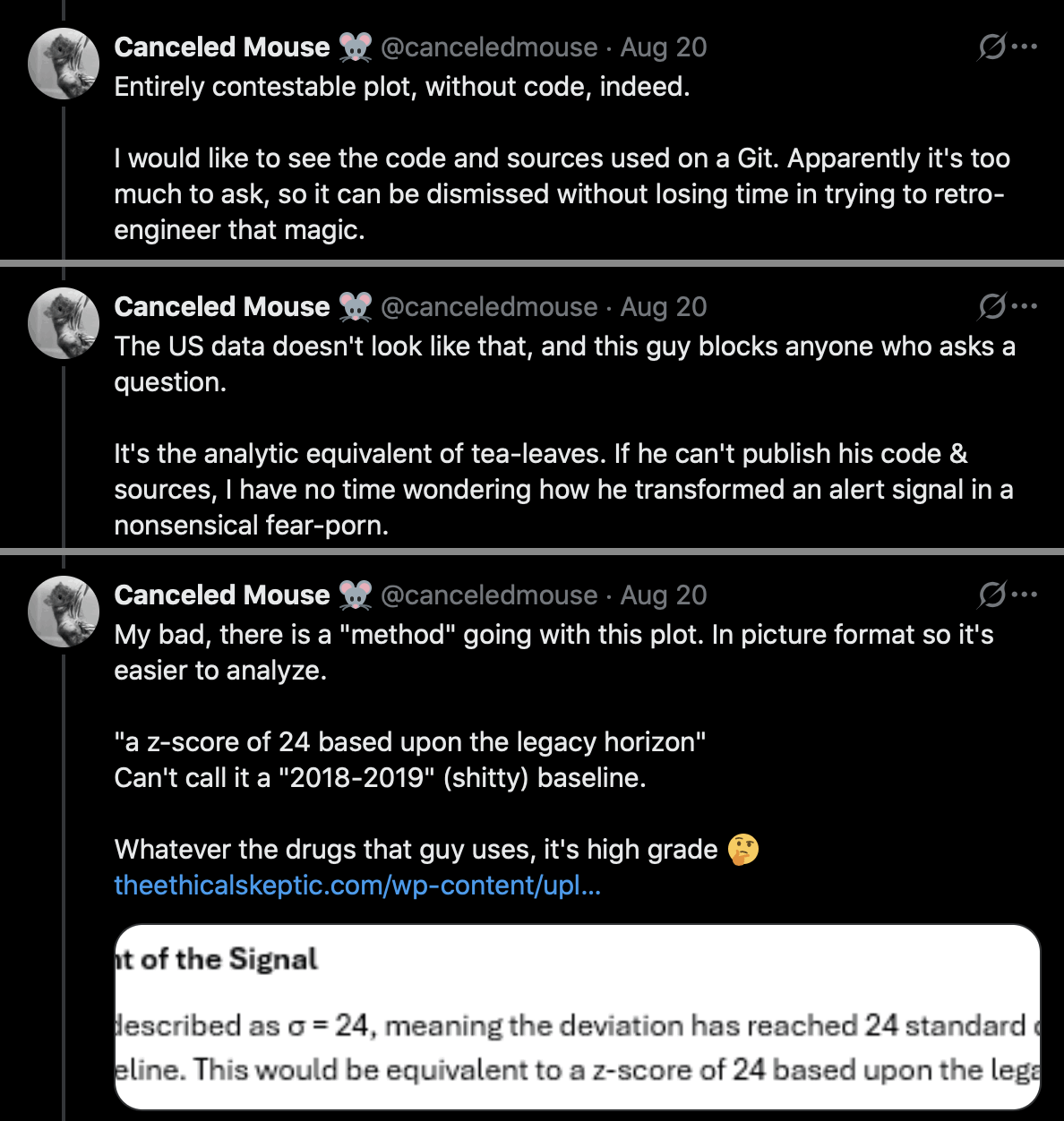
And canceledmouse said that Ethical Skeptic's plot is "entirely contestable" and "nonsensical fear porn": [https://x.com/canceledmouse/status/1958015548685525389]
The next plot shows deaths per mid-year resident population estimates going back to 1968, because at first I didn't find data for births going back further than 1995:
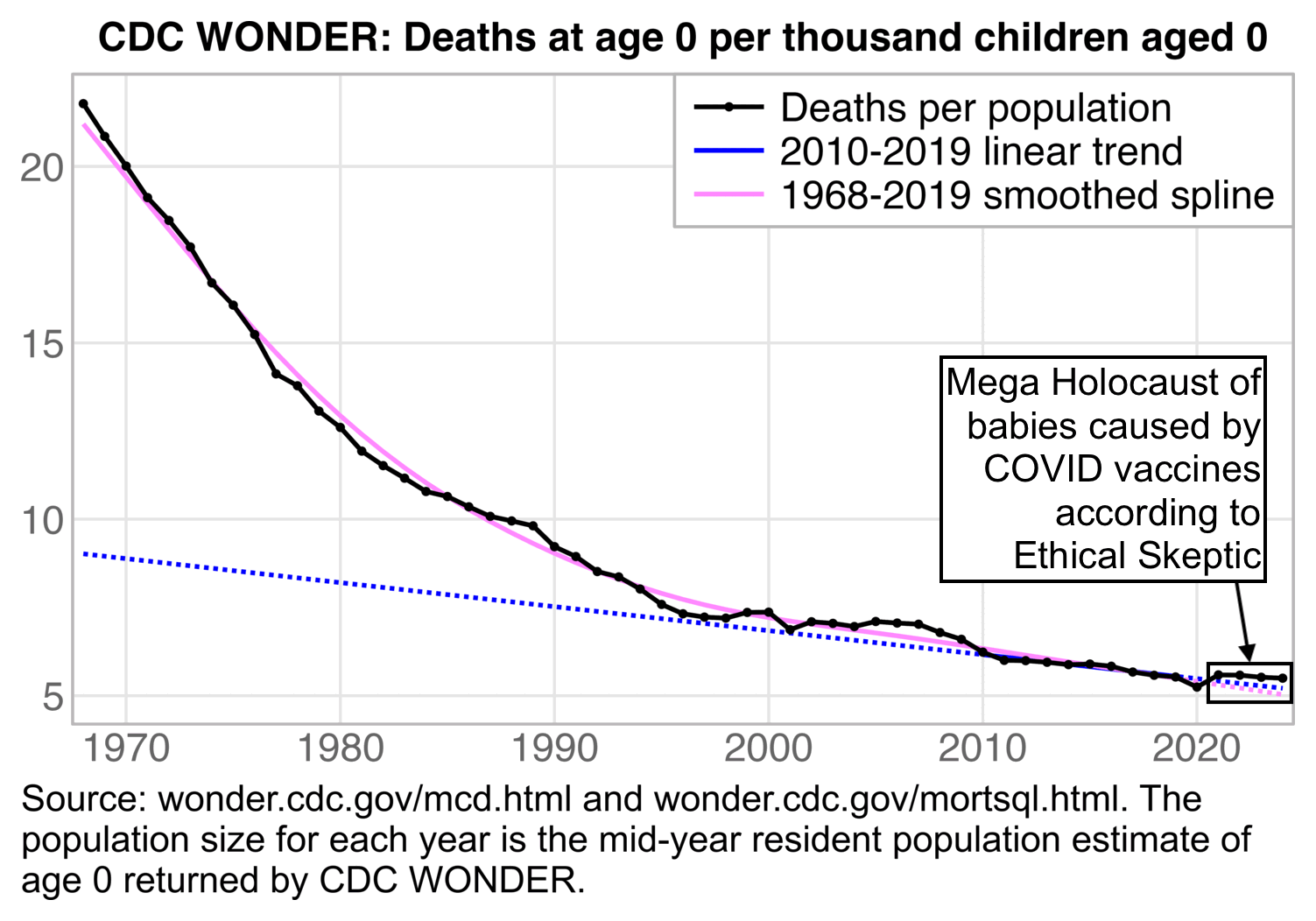
t=fread("https://sars2.net/f/wonderinfantdeadpop.csv")
p1=t[,.(x=year,y=dead/pop*1e3,z=1,fit=T)]
p2=copy(p1)[,z:=2][,fit:=x%in%2015:2019]
p2$y=predict(lm(y~x,p2[fit==T]),p2)
p3=copy(p1)[,z:=3][,fit:=x<2020]
p3$y=p3[fit==T,predict(smooth.spline(x,y,spar=.8),p3$x)]$y
p=rbind(p1,p2,p3)
p[,z:=factor(z,,c("Deaths per population","2015-2019 linear trend","1968-2019 smoothed spline"))]
xstart=1968-.5;xend=2024+.5;xbreak=seq(1970,xend,10)
ylim=p[,{x=extendrange(y);.(min=x[1],max=x[2])}]
ggplot(p)+
geom_rect(data=ylim,aes(ymin=min,ymax=max),xmin=xstart,xmax=xend,lineend="square",linejoin="mitre",fill=NA,color="gray72",linewidth=.4)+
geom_line(aes(x,y,color=z),linetype="11",linewidth=.6)+
geom_line(data=p[fit==T],aes(x,y,color=z),linewidth=.6)+
geom_line(data=p[z==z[1]],aes(x,y,color=z),linewidth=.6)+
geom_point(aes(x,y,color=z,alpha=z),stroke=0,size=1.2)+
labs(title="CDC WONDER: Deaths at age 0 per thousand children aged 0",x=NULL,y=NULL)+
scale_x_continuous(limits=c(xstart,xend),breaks=xbreak)+
scale_color_manual(values=c("black","blue","#ff88ff"))+
scale_alpha_manual(values=c(1,0,0))+
coord_cartesian(clip="off",expand=F)+
theme(axis.text=element_text(size=11,color="gray40",margin=margin(2,2,2,2)),
axis.ticks.length=unit(0,"pt"),
legend.background=element_rect(color="gray70",linewidth=.4),
legend.box.spacing=unit(0,"pt"),
legend.direction="vertical",
legend.justification=c(1,1),
legend.key=element_blank(),
legend.key.height=unit(12,"pt"),
legend.key.width=unit(24,"pt"),
legend.margin=margin(3,5,3,3),
legend.position=c(1,1),
legend.spacing.x=unit(2,"pt"),
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=11,vjust=.5),
legend.title=element_blank(),
panel.background=element_blank(),
panel.grid=element_blank(),
panel.grid.major=element_line(linewidth=.4,color="gray90"),
plot.subtitle=element_text(hjust=.5,margin=margin(,,4)),
plot.title=element_text(size=11,hjust=.5,face="bold",margin=margin(,,4)))
ggsave("1.png",width=5,height=3,dpi=300*4)
When I showed the plot above to Clare Craig, she posted this tweet: [https://x.com/ClareCraigPath/status/1960621848384651566]
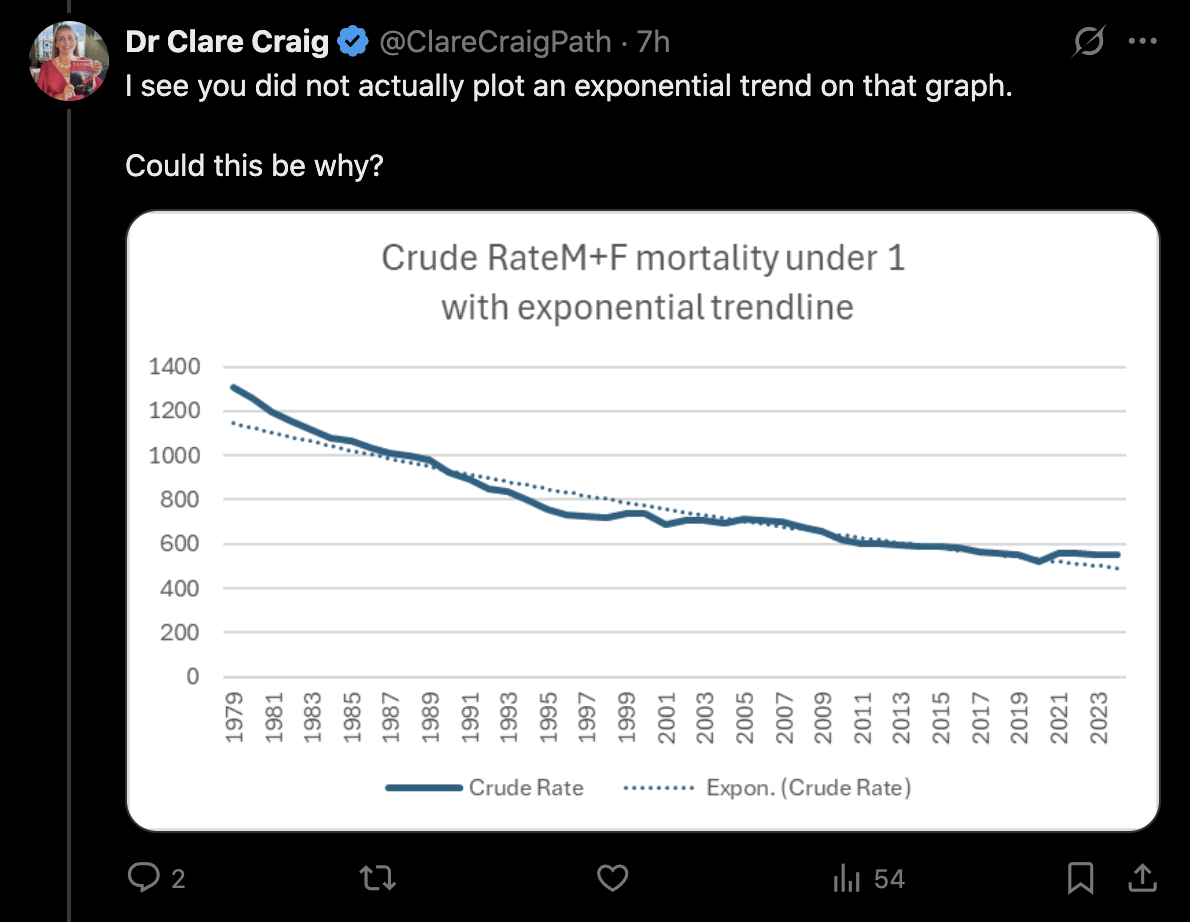
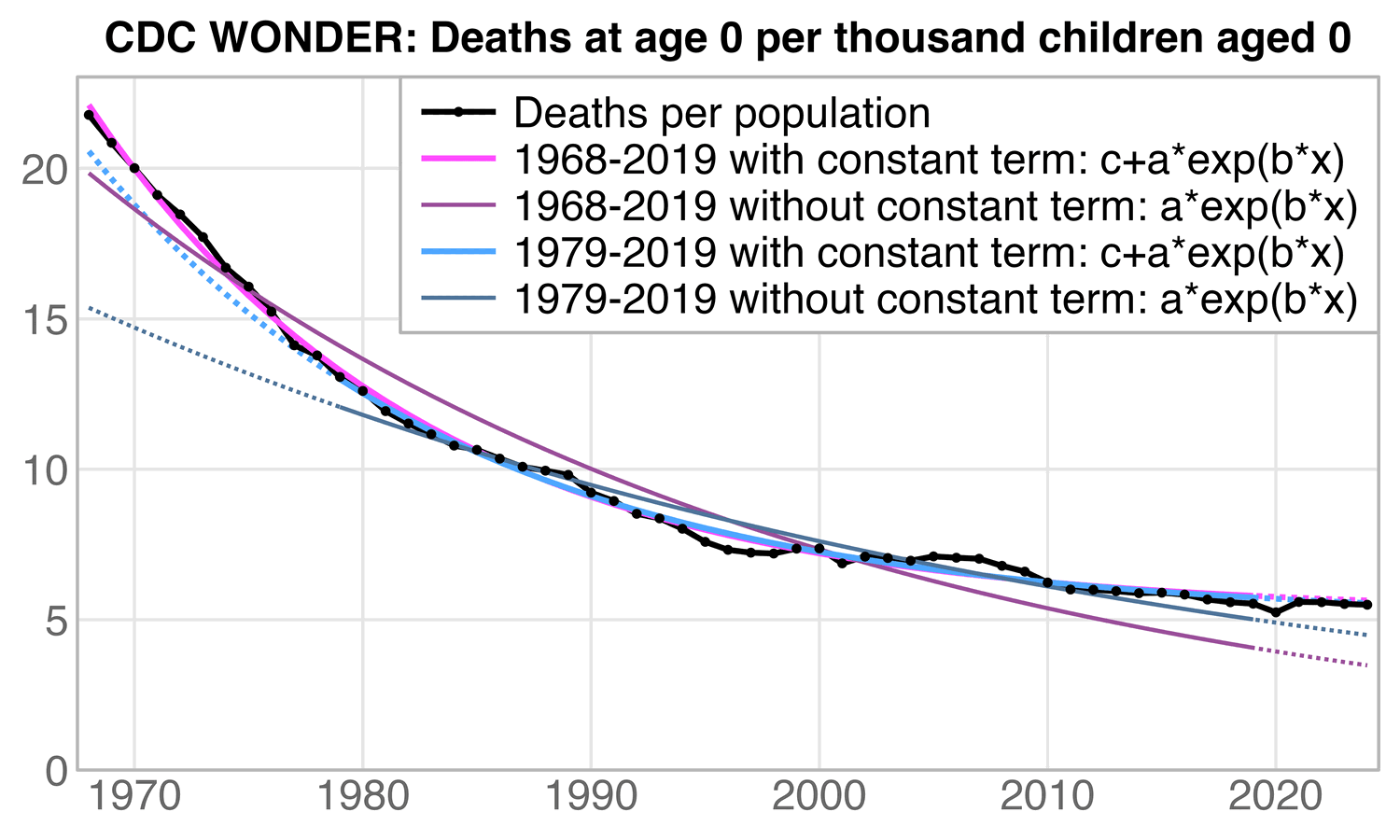
In fact she was right that I should've fitted an exponential function
to the data. But she made the mistake of leaving out a constant term
from her exponential function, so her function was of the form
a*exp(b*x) and not c+a*exp(b*x). When I added
a constant term, the mortality rate in 2021-2024 was below the
exponential trend regardless of whether I started the fitting period
from 1968 or from 1979 like her. I treated the first year of the fitting
period as year zero when I fitted the exponential functions:
t=fread("https://sars2.net/f/wonderinfantdeadpop.csv")
p1=t[,.(x=year,y=dead/pop*1e3,z=1,fit=T)]
p2=copy(p1)[,z:=2][,fit:=x<2020]
p3=copy(p1)[,z:=4][,fit:=x%in%1979:2019]
exp=rbind(p2,p3)
exp1=exp[,.(x,y=predict(nls(y~c+a*exp(b*(x-.SD[fit==T,x][1])),.SD[fit==T],.SD[fit==T,.(a=y[.N]-y[1],b=-.08,c=min(y))]),.SD),fit),z]
exp2=exp[,.(x,y=predict(nls(y~a*exp(b*(x-.SD[fit==T,x][1])),.SD[fit==T],.SD[fit==T,.(a=y[1],b=-.08)]),.SD),fit),z]
p=rbind(p1,exp1,copy(exp2)[,z:=z+1])
p[,z:=factor(z,,c("Deaths per population","1968-2019 with constant term: c+a*exp(b*x)","1968-2019 without constant term: a*exp(b*x)","1979-2019 with constant term: c+a*exp(b*x)","1979-2019 without constant term: a*exp(b*x)"))]
xstart=1968-.5;xend=2024+.5;xbreak=seq(1970,xend,10)
ylim=p[,{x=extendrange(y);.(min=0,max=x[2])}]
ggplot(p)+
geom_rect(data=ylim,aes(ymin=min,ymax=max),xmin=xstart,xmax=xend,lineend="square",linejoin="mitre",fill=NA,color="gray72",linewidth=.4)+
geom_line(aes(x,y,color=z,linewidth=z),linetype="11")+
geom_line(data=p[fit==T],aes(x,y,color=z,linewidth=z))+
geom_point(aes(x,y,color=z,alpha=z),stroke=0,size=1.2)+
labs(title="CDC WONDER: Deaths at age 0 per thousand children aged 0",x=NULL,y=NULL)+
scale_x_continuous(limits=c(xstart,xend),breaks=xbreak)+
scale_color_manual(values=c("black",hsv(c(30,30,21,21)/36,c(.7,.5,.7,.5),c(1,.6,1,.6))))+
scale_alpha_manual(values=c(1,0,0,0,0))+
scale_linewidth_manual(values=c(.7,.7,.5,.7,.5))+
coord_cartesian(clip="off",expand=F)+
theme(axis.text=element_text(size=11,color="gray40",margin=margin(2,2,2,2)),
axis.ticks.length=unit(0,"pt"),
legend.background=element_rect(color="gray70",linewidth=.4),
legend.box.spacing=unit(0,"pt"),
legend.direction="vertical",
legend.justification=c(1,1),
legend.key=element_blank(),
legend.key.height=unit(12,"pt"),
legend.key.width=unit(24,"pt"),
legend.margin=margin(3,5,3,3),
legend.position=c(1,1),
legend.spacing.x=unit(2,"pt"),
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=11,vjust=.5),
legend.title=element_blank(),
panel.background=element_blank(),
panel.grid=element_blank(),
panel.grid.major=element_line(linewidth=.4,color="gray90"),
plot.subtitle=element_text(hjust=.5,margin=margin(,,4)),
plot.title=element_text(size=11,hjust=.5,face="bold",margin=margin(,,4)))
ggsave("1.png",width=5,height=3,dpi=300*4)
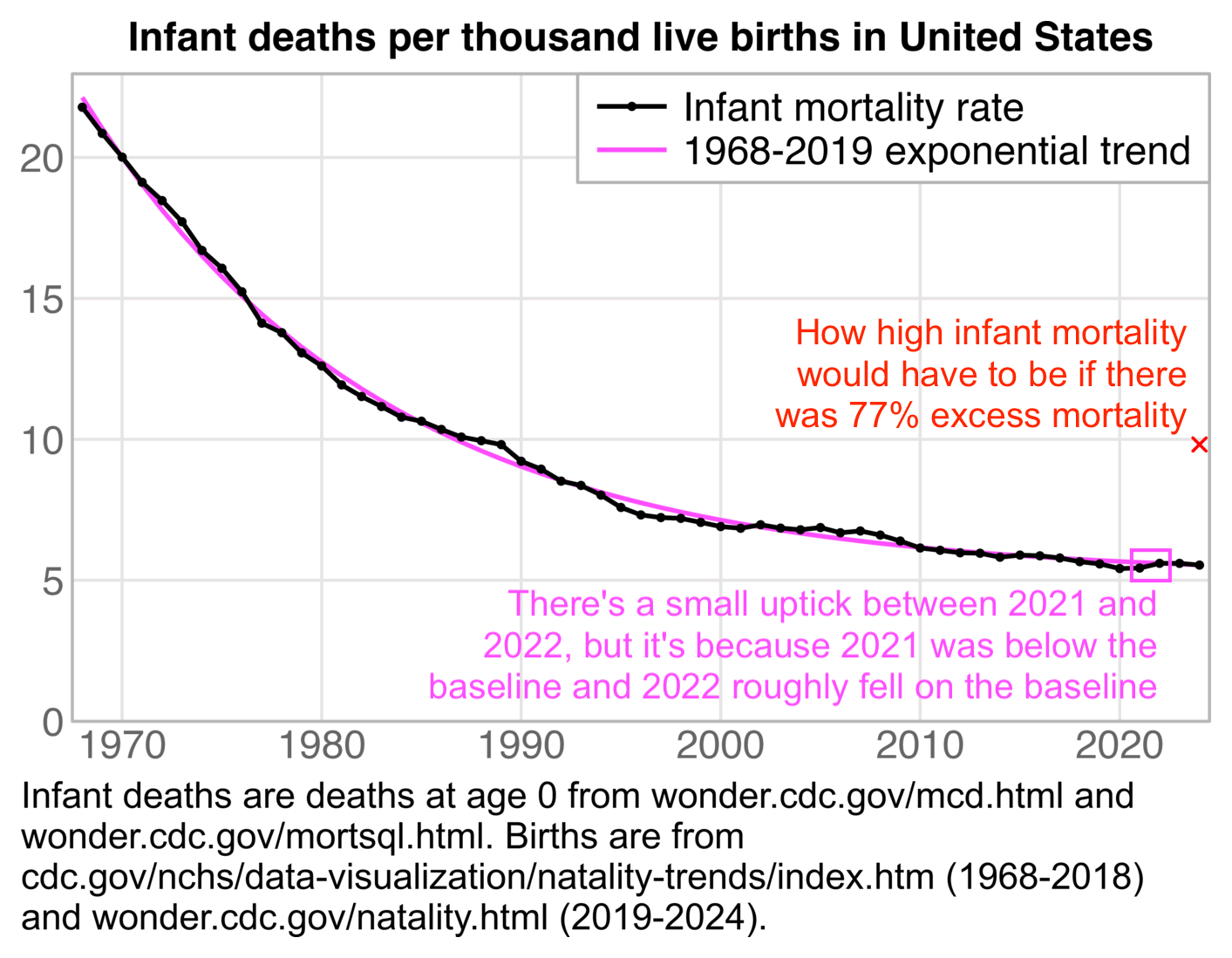
Next I found data for births going back to 1909. [https://cdc.gov/nchs/data-visualization/natality-trends/index.htm] When I plotted the yearly ratio of deaths per live births since 1968, and I fitted an exponential function to the ratio, I got about -4% excess infant mortality in 2020, -4% in 2021, 0% in 2022, 0% in 2023, and 0% in 2024:
t=CJ(year=1968:2024)
t$born=c(3501564,3600206,3731386,3555970,3258411,3136965,3159958,3144198,3167788,3326632,3333279,3494398,3612258,3629238,3680537,3638933,3669141,3760561,3756547,3809394,3909510,4040958,4158212,4110907,4065014,4000240,3952767,3899589,3891494,3880894,3941553,3959417,4058814,4025933,4021726,4089950,4112052,4138349,4265555,4316233,4247694,4130665,3999386,3953590,3952841,3932181,3988076,3978497,3945875,3855500,3791712,3747540,3613647,3664292,3667758,3596017,3618267)
t=merge(fread("https://sars2.net/f/wonderinfantdeadpop.csv")[,.(year,dead)],t)
p1=t[,.(x=year,y=dead/born*1e3,z=1,fit=T)]
p2=copy(p1)[,z:=2][,fit:=x<2020]
p2=p2[,.(x,y=predict(nls(y~c+a*exp(b*(x-.SD[fit==T,x][1])),.SD[fit==T],.SD[fit==T,.(a=y[.N]-y[1],b=-.08,c=min(y))]),.SD),fit,z=2)]
p=rbind(p1,p2)
p[,z:=factor(z,,c("Infant mortality rate","1968-2019 exponential trend"))]
xstart=1968-.5;xend=2024+.5;xbreak=seq(1970,xend,10)
ylim=p[,{x=extendrange(y);.(min=0,max=x[2])}]
ggplot(p)+
geom_rect(data=ylim,aes(ymin=min,ymax=max),xmin=xstart,xmax=xend,lineend="square",linejoin="mitre",fill=NA,color="gray72",linewidth=.4)+
geom_line(aes(x,y,color=z),linewidth=.6)+
geom_point(aes(x,y,color=z,alpha=z),stroke=0,size=1.2)+
geom_line(data=p[z==z[1]],aes(x,y,color=z),linewidth=.6)+
labs(title="CDC WONDER: Infant deaths per thousand live births",x=NULL,y=NULL)+
scale_x_continuous(limits=c(xstart,xend),breaks=xbreak)+
scale_color_manual(values=c("black",hsv(5/6,.7,1)))+
scale_alpha_manual(values=1:0)+
coord_cartesian(clip="off",expand=F)+
theme(axis.text=element_text(size=11,color="gray40",margin=margin(2,2,2,2)),
axis.ticks.length=unit(0,"pt"),
legend.background=element_rect(color="gray70",linewidth=.4),
legend.box.spacing=unit(0,"pt"),
legend.direction="vertical",
legend.justification=c(1,1),
legend.key=element_blank(),
legend.key.height=unit(12,"pt"),
legend.key.width=unit(24,"pt"),
legend.margin=margin(3,5,3,3),
legend.position=c(1,1),
legend.spacing.x=unit(2,"pt"),
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=11,vjust=.5),
legend.title=element_blank(),
panel.background=element_blank(),
panel.grid=element_blank(),
panel.grid.major=element_line(linewidth=.4,color="gray90"),
plot.subtitle=element_text(hjust=.5,margin=margin(,,4)),
plot.title=element_text(size=11,hjust=.5,face="bold",margin=margin(,,4)))
ggsave("1.png",width=4.7,height=3,dpi=300*4)
The x-axis in Ethical Skeptic's plots starts from 2018, so it's hard to tell if the increase in deaths between 2021 and 2022 was because there was low mortality in 2021 but normal mortality in 2022, or because there was normal mortality in 2021 but high mortality in 2022, or because there was high mortality in 2021 but even higher mortality in 2022. It's one of many reasons why he should plot data going back further than 2018.
Here among women aged 15-49, the percentage increase in drug overdose deaths between 2018 and 2023 was about 166% in American Indians, 144% in blacks, 87% in Hispanics, 50% in Asians and Pacific Islanders, and 16% in whites:
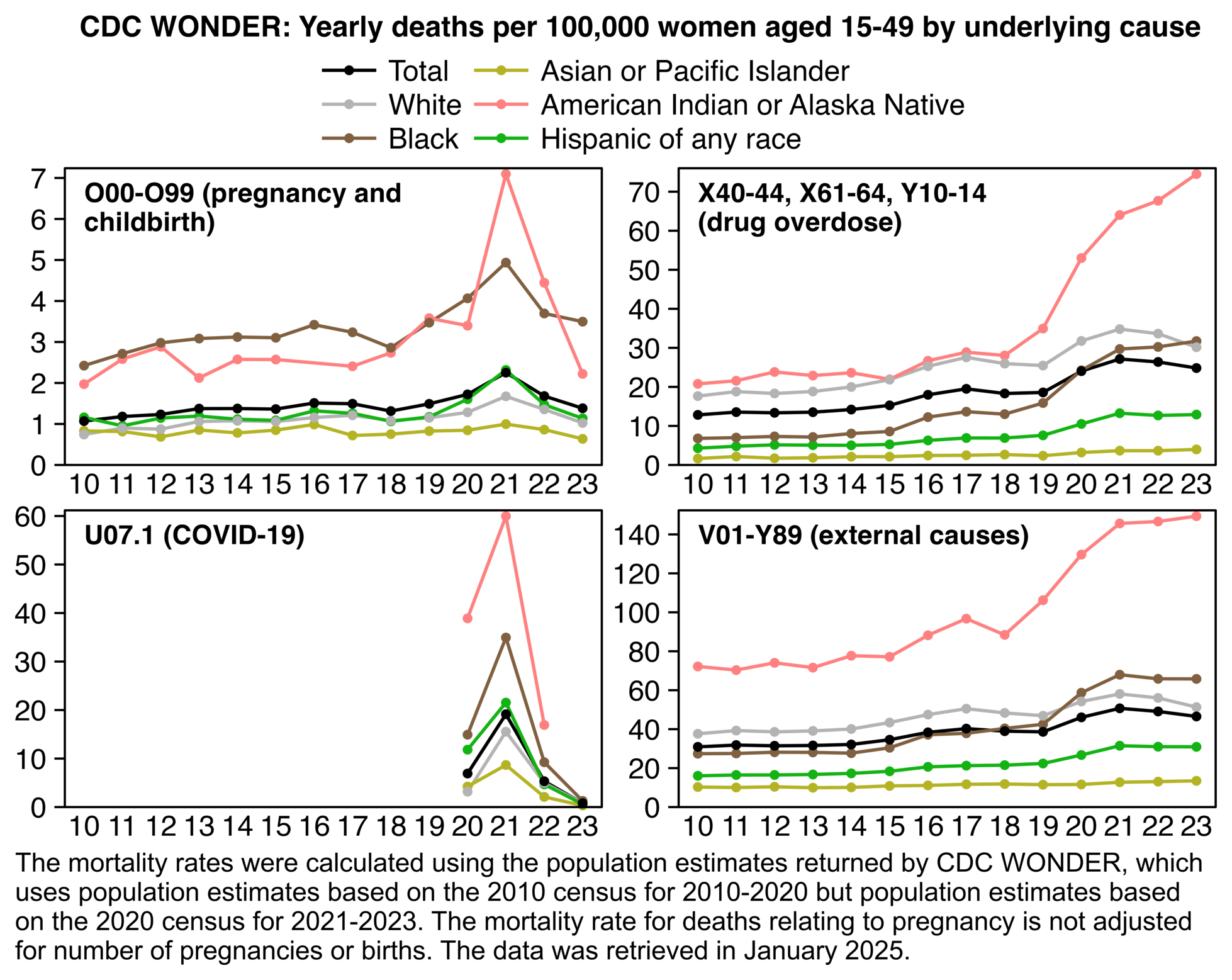
t=fread("https://sars2.net/f/wonderpregnancyrace.csv")
t[race%like%"Asian|Pacific",race:="Asian or Pacific Islander"]
t[race=="Black or African American",race:="Black"]
t[cause=="O00-O99 (Pregnancy, childbirth and the puerperium)",cause:="O00-O99 (pregnancy and childbirth)"]
t[,cause:=factor(cause,unique(cause))]
t[hispanic=="Hispanic or Latino",race:="Hispanic of any race"]
t[,hispanic:=NULL]
t=rbind(t,t[,.(dead=sum(dead,na.rm=T),pop=sum(pop,na.rm=T),race="Total"),.(cause,year)])
t=t[race!="More than one race"]
t[,race:=factor(race,c("Total","White","Black","Asian or Pacific Islander","American Indian or Alaska Native","Hispanic of any race"))]
p=t[,.(y=sum(dead,na.rm=T)/sum(pop,na.rm=T)*1e5),.(x=year,cause,z=race)]
xstart=2010;xend=2023;xbreak=xstart:xend
p=p[x%in%xstart:xend]
ylim=c(0,max(p$y)*1.02);ybreak=pretty(ylim,7)
ggplot(p,aes(x,y,z))+
facet_wrap(~cause,ncol=2,strip.position="top",dir="v",scales="free")+
geom_line(aes(color=z),linewidth=.6)+
geom_point(aes(color=z),size=1.1)+
geom_text(data=p[,max(y),cause],aes(label=stringr::str_wrap(cause,26),y=V1*.97),x=xstart,fontface=2,lineheight=.9,hjust=0,vjust=1,size=3.6)+
labs(x=NULL,y=NULL,title="CDC WONDER: Yearly deaths per 100,000 women aged 15-49 by underlying cause")+
scale_x_continuous(limits=c(xstart-.5,xend+.5),breaks=xbreak,labels=substr(xbreak,3,4))+
scale_y_continuous(limits=\(x)c(0,max(x)*1.02),breaks=\(x)pretty(x,7))+
scale_color_manual(values=c("black","gray70",hsv(3/36,.5,.5),hsv(1/6,.8,.7),hsv(0,.5,1),hsv(12/36,.9,.7)))+
coord_cartesian(clip="off",expand=F)+
guides(color=guide_legend(ncol=2,byrow=F))+
theme(axis.text=element_text(size=11,color="black"),
axis.text.x=element_text(margin=margin(3)),
axis.text.y=element_text(margin=margin(,3)),
axis.ticks=element_line(linewidth=.4,color="black"),
axis.ticks.length.x=unit(0,"pt"),
axis.ticks.length.y=unit(4,"pt"),
legend.background=element_blank(),
legend.box.background=element_blank(),
legend.box.spacing=unit(0,"pt"),
legend.direction="vertical",
legend.justification="center",
legend.key=element_blank(),
legend.key.height=unit(13,"pt"),
legend.key.width=unit(26,"pt"),
legend.margin=margin(,,5),
legend.position="top",
legend.spacing.x=unit(2,"pt"),
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=11,vjust=.5),
legend.title=element_blank(),
panel.background=element_blank(),
panel.border=element_rect(fill=NA,linewidth=.4),
panel.spacing=unit(4,"pt"),
plot.margin=margin(5,5,2,5),
plot.title=element_text(size=11,hjust=.5,face=2,margin=margin(2,,4)),
strip.background=element_blank(),
strip.text=element_blank())
ggsave("1.png",width=6.5,height=4.5,dpi=300*4)
But here the percentage change in infant mortality between 2018 and 2023 was about 13% in Native Americans, 3% in Hispanics, 3% in blacks, -4% in Asians and Pacific Islanders, and -4% in whites. So Native Americans had the biggest increase in both infant mortality and drug deaths:
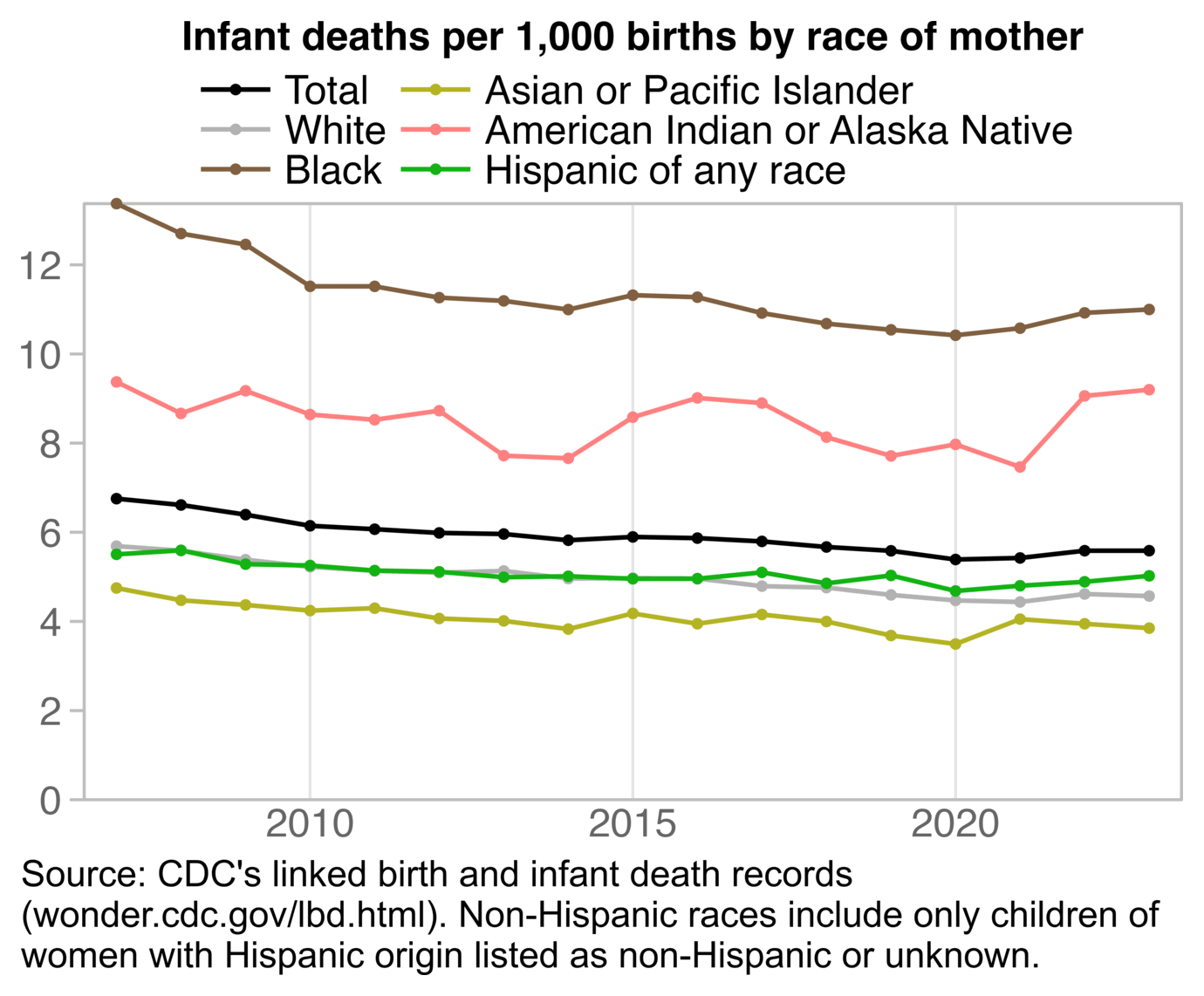
t=fread("https://sars2.net/f/wonderlinkedinfantrace.csv")
t2=t[!hispanic%like%"Non-Hispanic|unknown"][,race:="Hispanic of any race"]
t=rbind(t[hispanic%like%"Non-Hispanic|unknown"],t2)
t=t[!race%like%"Not Reported|More than one race"]
t[race=="Black or African American",race:="Black"]
t[race%like%"Asian|Pacific",race:="Asian or Pacific Islander"]
t[,hispanic:=NULL]
t=rbind(t,copy(t)[,race:="Total"])
t=t[,.(dead=sum(dead),born=sum(born)),.(race,year)]
p=t[,.(y=sum(dead)/sum(born)*1000),.(x=year,z=race)]
p[,z:=factor(z,strsplit("Total
White
Black
Asian or Pacific Islander
American Indian or Alaska Native
Hispanic of any race","\\n")[[1]])]
xstart=2006.5;xend=2023.5;xbreak=seq(2000,xend-1,5)
p=p[x>=xstart&x<=xend]
ylim=p[,.(min=0,max=max(y))]
ggplot(p)+
geom_vline(xintercept=xbreak,color="gray90",linewidth=.4)+
geom_rect(data=ylim,aes(ymin=min,ymax=max),xmin=xstart,xmax=xend,lineend="square",linejoin="mitre",fill=NA,color="gray75",linewidth=.4)+
geom_line(aes(x,y,color=z),linewidth=.6)+
geom_point(aes(x,y,color=z),stroke=0,size=1.5)+
labs(title="Infant deaths per 1,000 births by race of mother",x=NULL,y=NULL)+
scale_x_continuous(limits=c(xstart,xend),breaks=xbreak)+
scale_y_continuous(breaks=pretty)+
scale_color_manual(values=c("black","gray70",hsv(3/36,.5,.5),hsv(1/6,.8,.7),hsv(0,.5,1),hsv(12/36,.9,.7)))+
coord_cartesian(clip="off",expand=F)+
guides(color=guide_legend(ncol=2,byrow=F))+
theme(axis.text=element_text(size=11,color="gray40",margin=margin(2,2,2,2)),
axis.ticks=element_line(linewidth=.4,color="gray75"),
axis.ticks.length.x=unit(0,"pt"),
axis.ticks.length.y=unit(4,"pt"),
legend.background=element_blank(),
legend.box.spacing=unit(0,"pt"),
legend.direction="vertical",
legend.key=element_blank(),
legend.key.height=unit(11,"pt"),
legend.key.width=unit(24,"pt"),
legend.margin=margin(,,4),
legend.position="top",
legend.spacing.x=unit(1.5,"pt"),
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=11,vjust=.5),
legend.title=element_blank(),
panel.background=element_blank(),
panel.grid=element_blank(),
panel.spacing=unit(3,"pt"),
plot.title=element_text(size=11,hjust=.5,face="bold",margin=margin(,,3)),
strip.background=element_blank(),
strip.text=element_blank())
ggsave("1.png",width=4.6,height=3.3,dpi=300*4)
When I showed Kevin McCairn that I got negative excess infant mortality with the 1968-2019 exponential baseline, he argued that my analysis was trumped my Clare Craig's plots, where she relied on a 2015-2019 linear baseline to suggest that vaccines had caused excess infant deaths in 2021-2024: [https://x.com/KevinMcCairnPhD/status/1960934734915363052, https://drclarecraig.substack.com/p/battle-of-the-baselines]
But I showed McCairn that a 2015-2019 linear baseline produced positive excess births each year between 2021 and 2024. So did the vaccines cause a baby boom?
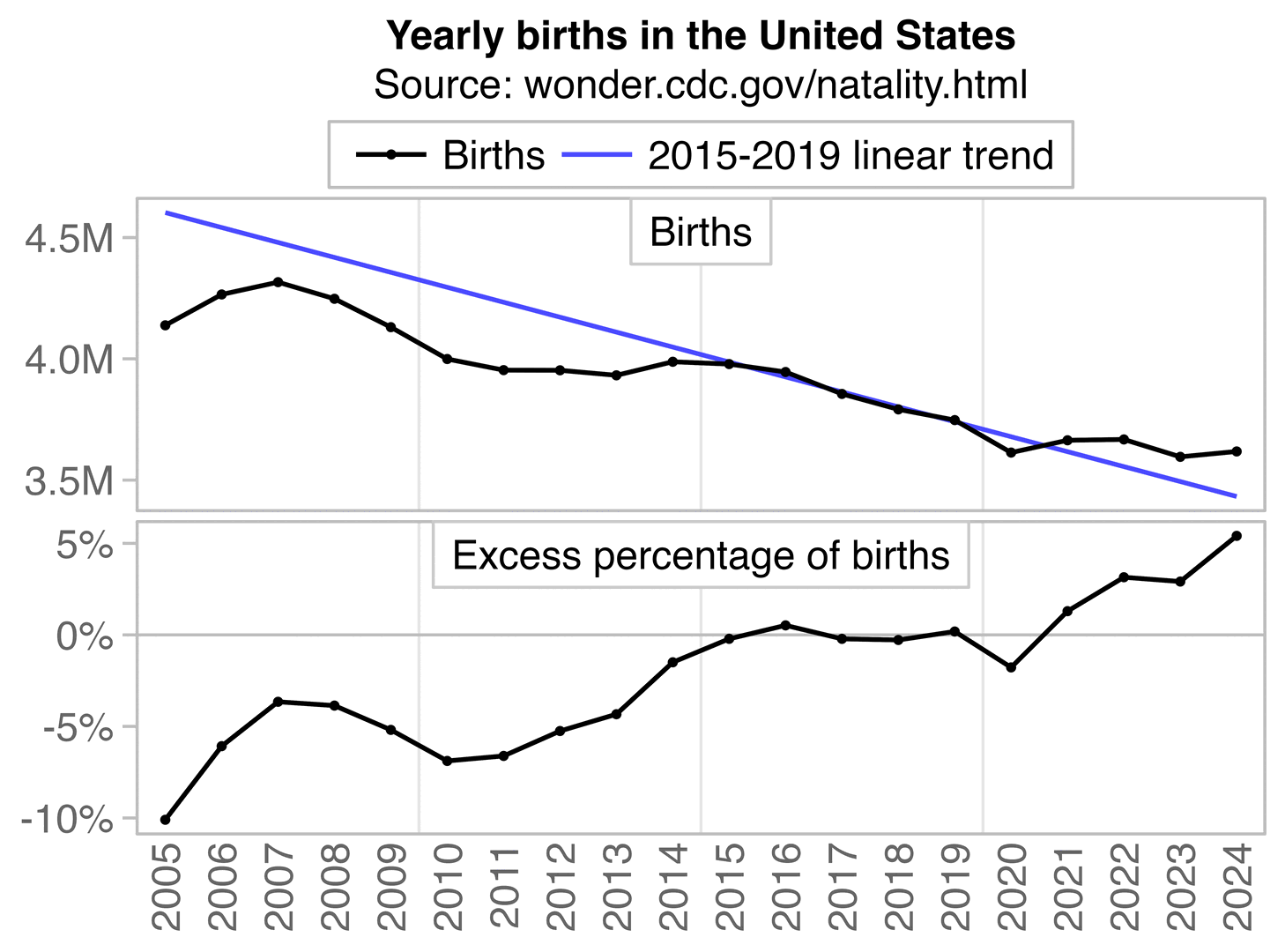
Clare Craig pointed out that the percentage of infant deaths in females out of total infant deaths had increased since 2021: [https://drclarecraig.substack.com/p/battle-of-the-baselines]
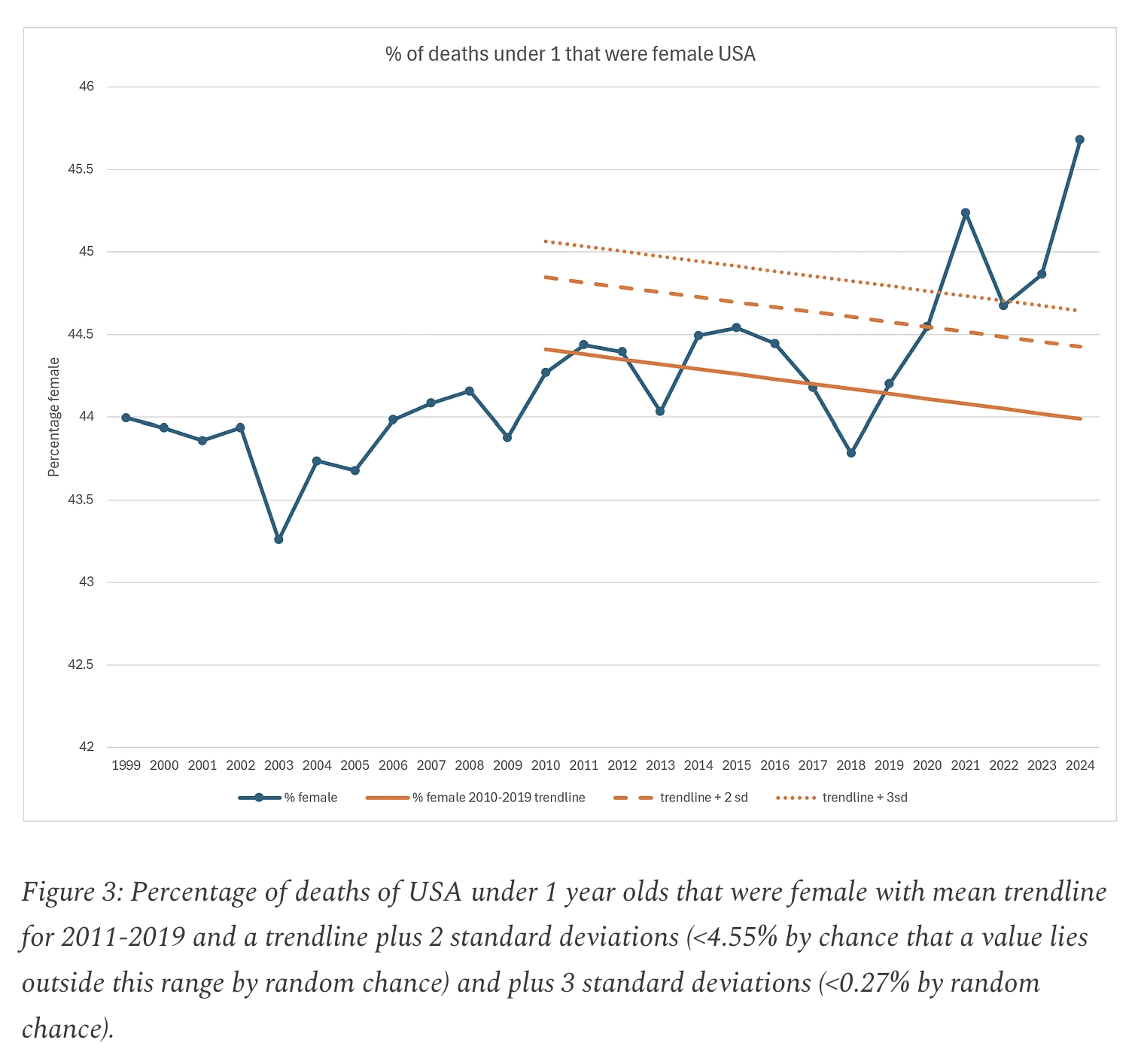
However there's been an increasing trend in the percentage since at least the 1970s. The ratio fell close to the long-term trend in 2022 and 2023, even though by several measures excess infant mortality was higher in 2022 and 2023 than 2021. There wasn't a clear increase in the ratio above the long-term trend until 2024-2025, but by then the effect of COVID vaccines should've presumably been weaker than in 2021-2023:
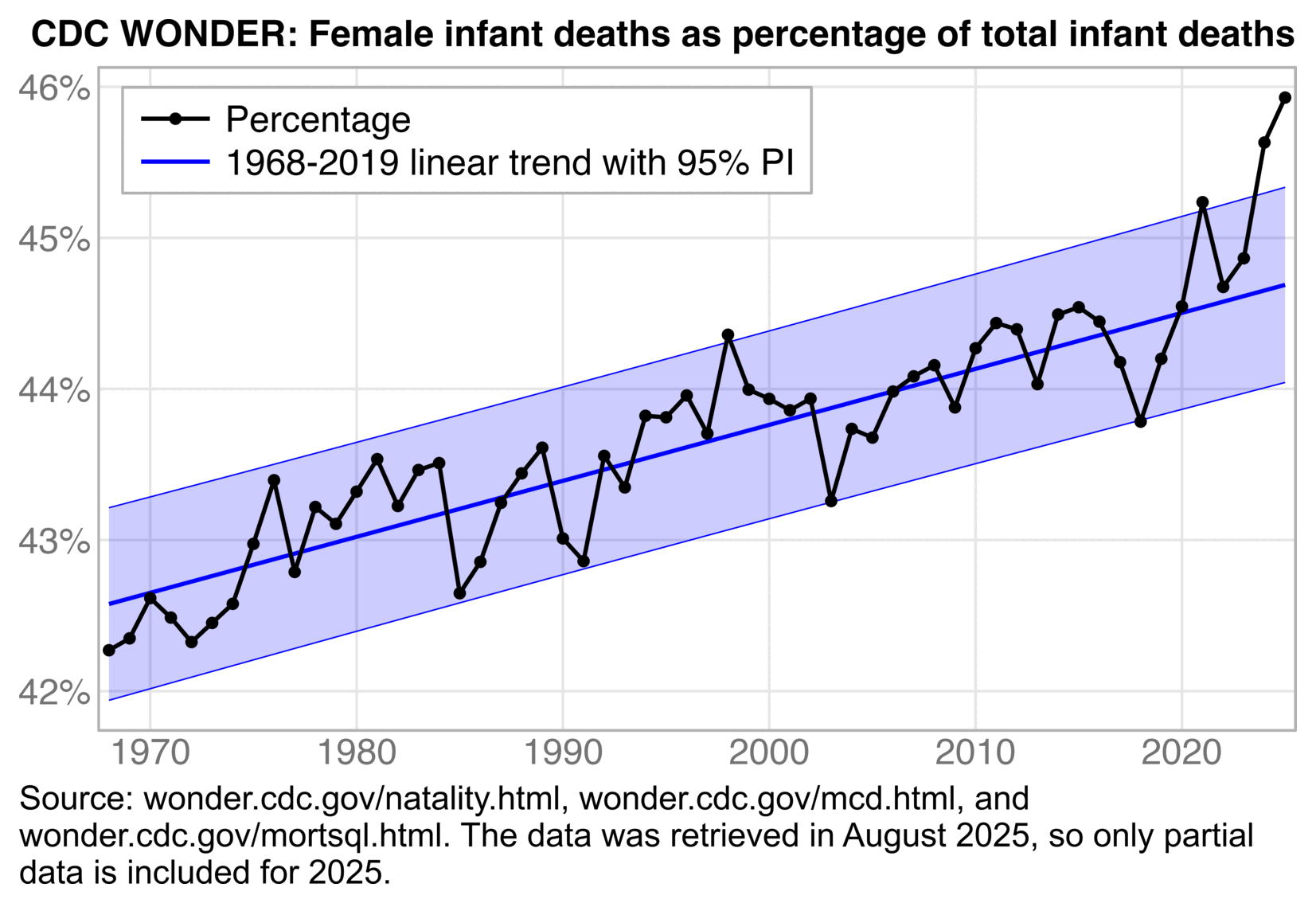
t=fread("https://sars2.net/f/wonderinfantsexyearly.csv")
p=t[,.(y=dead[sex=="F"]/sum(dead)*100),.(x=year)]
pred=p[,.(x,predict(lm(y~x,.SD[x<2020]),.SD,interval="prediction"))]
p=p[,.(x,y=c(y,pred$fit),z=rep(1:2,each=.N))]
p[,z:=factor(z,,c("Percentage","1968-2019 linear trend with 95% PI"))]
xstart=1968-.5;xend=2025+.5;xbreak=seq(1970,xend,10)
ylim=extendrange(c(p$y,pred$lwr,pred$upr));ybreak=0:100
ggplot(p)+
geom_hline(yintercept=0,color="gray70",linewidth=.4)+
geom_ribbon(data=pred,aes(x,ymin=lwr,ymax=upr),fill="blue",color="blue",alpha=.2,linewidth=.2)+
annotate("rect",xmin=xstart,xmax=xend,ymin=ylim[1],ymax=ylim[2],linewidth=.4,lineend="square",linejoin="mitre",fill=NA,color="gray70")+
geom_line(aes(x,y,color=z),linewidth=.6)+
geom_point(aes(x,y,alpha=z),size=1.1)+
geom_line(data=p[z==z[1]],aes(x,y,color=z),linewidth=.6)+
labs(x=NULL,y=NULL,title="CDC WONDER: Female infant deaths as percentage of total infant deaths")+
scale_x_continuous(limits=c(xstart,xend),breaks=xbreak)+
scale_y_continuous(limits=ylim,breaks=ybreak,labels=\(x)paste0(x,"%"))+
scale_color_manual(values=c("black","blue"))+
scale_alpha_manual(values=1:0)+
coord_cartesian(clip="off",expand=F)+
theme(axis.text=element_text(size=11,color="gray45"),
axis.ticks=element_line(linewidth=.4,color="black"),
axis.ticks.length=unit(0,"pt"),
legend.background=element_rect(color="gray70",linewidth=.4),
legend.box.spacing=unit(0,"pt"),
legend.justification=c(0,1),
legend.key=element_blank(),
legend.key.height=unit(13,"pt"),
legend.key.width=unit(26,"pt"),
legend.margin=margin(3,5,3,3),
legend.position=c(.02,.97),
legend.spacing.x=unit(2,"pt"),
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=11),
legend.title=element_blank(),
panel.background=element_blank(),
panel.grid.major=element_line(linewidth=.4,color="gray90"),
plot.title=element_text(size=11,hjust=1,face=2,margin=margin(,,4)))
ggsave("1.png",width=5.5,height=3.3,dpi=300*4)
Kevin McCairn said that I should use a negative binomial general additive model instead of a linear baseline: [https://x.com/KevinMcCairnPhD/status/1960826535629423073]
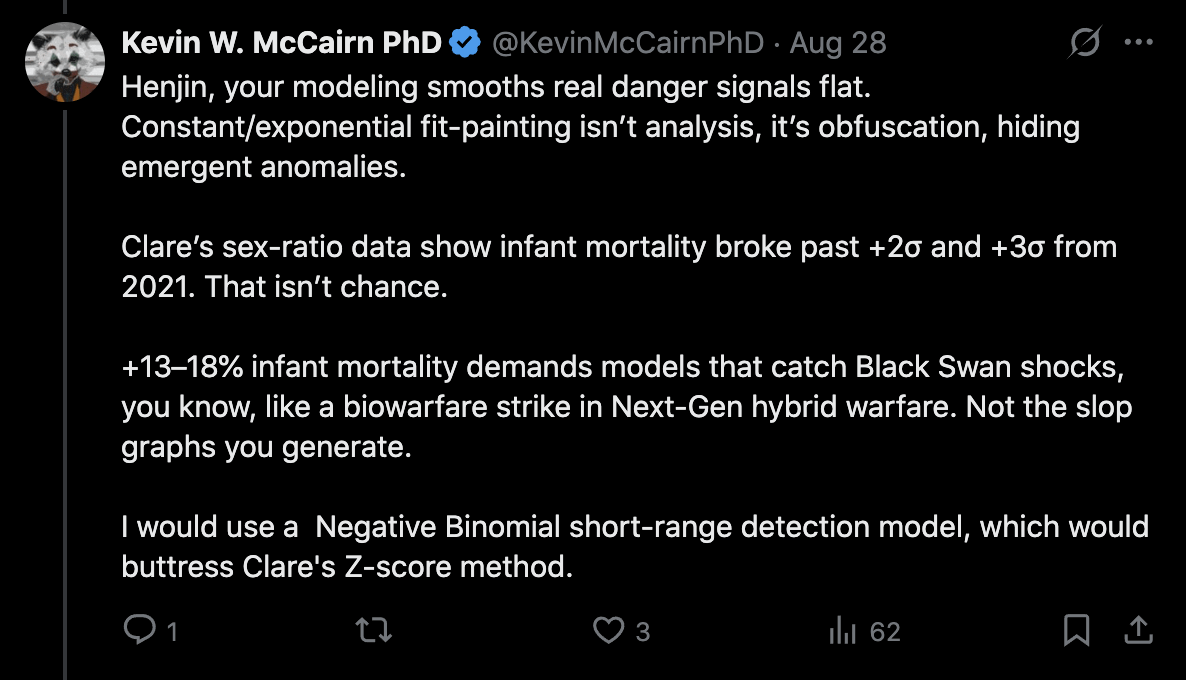
In the next plot which shows infant mortality rate by sex, I used the
mgcv::gam function to fit a negative binomial GAM to the
mortality in 2004-2019. In December 2024 which was a moving average for
the entire year of 2024, I got about 4% excess mortality in females and
-5% in males:
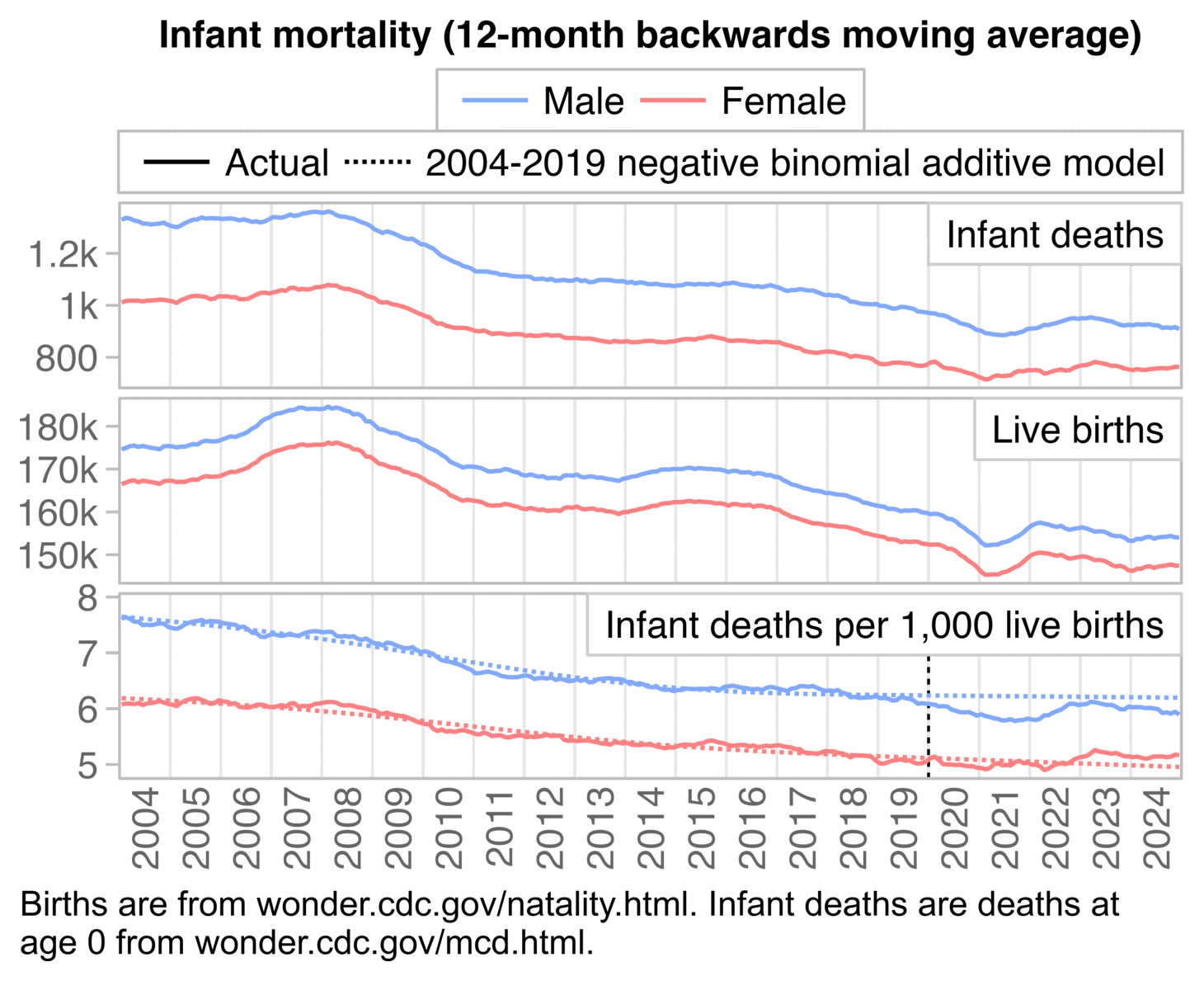
library(mgcv)
t=fread("https://sars2.net/f/wonderinfantsexmonthly.csv")
t=t[,.(x=rleid(year,month),year,month,dead=ma(dead,11,0),born=ma(born,11,0)),sex]
lm=t[,.(x,base=as.double(predict(gam(dead~s(x,k=4)+offset(log(born)),nb(),.SD[year<2020],method="REML"),.SD,type="response"))),sex]
t=merge(t,lm)
p=t[,.(sex,x=as.Date(paste(year,month,16,sep="-")),y=c(dead,born,c(dead,base)/born*1e3),z=rep(1:2,.N*c(3,1)),facet=rep(c(1:3,3),each=.N))]
p[,facet:=factor(facet,,c("Infant deaths","Live births","Infant deaths per 1,000 live births"))]
p[,z:=factor(z,,c("Actual","2004-2019 negative binomial additive model"))]
p[,sex:=factor(sex,c("M","F"),c("Male","Female"))]
xstart=as.Date("2004-1-1");xend=as.Date("2025-1-1");xbreak=seq(xstart+182,xend,"year")
p=p[x<=xend]
ylim=p[,{x=extendrange(y);.(min=x[1],max=x[2])},facet]
ggplot(p)+
facet_wrap(~facet,ncol=1,dir="v",scales="free_y")+
geom_vline(xintercept=seq(xstart,xend,"year"),color="gray90",linewidth=.4)+
geom_segment(data=ylim[3,.(facet,x=as.Date("2020-1-1"),min,max)],aes(x,min,xend=x,yend=max),linewidth=.4,linetype="22")+
geom_rect(data=ylim,aes(ymin=min,ymax=max),xmin=xstart,xmax=xend,lineend="square",linejoin="mitre",fill=NA,color="gray75",linewidth=.4)+
geom_line(aes(x,y,color=sex,linetype=z),linewidth=.6)+
geom_label(data=ylim,aes(xend,max,label=facet),vjust=1,hjust=1,label.r=unit(0,"pt"),label.padding=unit(5,"pt"),label.size=.4,size=3.87,fill="white",color="gray80")+
geom_label(data=ylim,aes(xend,max,label=facet),vjust=1,hjust=1,label.r=unit(0,"pt"),label.padding=unit(5,"pt"),label.size=0,size=3.87,fill=NA)+
labs(title="Infant mortality (12-month backwards moving average)",x=NULL,y=NULL)+
scale_x_continuous(limits=c(xstart,xend),breaks=xbreak,labels=year(xbreak))+
scale_y_continuous(breaks=\(x)pretty(x,4),labels=\(x)ifelse(x>=1e3,paste0(x/1e3,"k"),x))+
scale_color_manual(values=hsv(c(22,0)/36,.5,1))+
scale_linetype_manual(values=c("solid","11"))+
coord_cartesian(clip="off",expand=F)+
guides(color=guide_legend(order=1),linetype=guide_legend(order=2))+
theme(axis.text=element_text(size=11,color="gray40",margin=margin(2,2,2,2)),
axis.text.x=element_text(angle=90,vjust=.5,hjust=1),
axis.ticks=element_line(linewidth=.4,color="gray75"),
axis.ticks.length.x=unit(0,"pt"),
axis.ticks.length.y=unit(4,"pt"),
legend.background=element_rect(color="gray75",linewidth=.4),
legend.box="vertical",
legend.box.margin=margin(,,3),
legend.box.spacing=unit(0,"pt"),
legend.key=element_blank(),
legend.key.height=unit(12,"pt"),
legend.key.width=unit(24,"pt"),
legend.margin=margin(3,5,3,3),
legend.position="top",
legend.spacing.x=unit(2,"pt"),
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=11),
legend.title=element_blank(),
panel.background=element_blank(),
panel.grid=element_blank(),
panel.spacing=unit(3,"pt"),
plot.title=element_text(size=11,hjust=.5,face="bold",margin=margin(1,,4)),
strip.background=element_blank(),
strip.text=element_blank())
ggsave("1.png",width=4.86,height=3.6,dpi=300*4)
McCairn complained to me that my baseline was too coarse, and that I should've used a higher k-value in order to detect short-term fluctuations in the trend. He also kept saying that there was a reduction in birth rate caused by the vaccines. So I showed him the next plot where I plotted births instead of an infant mortality rate, where I got lower excess births since 2021 when I used a more coarse k-value of 2, but I got higher excess births since 2021 when I used a less coarse k-value of 8:
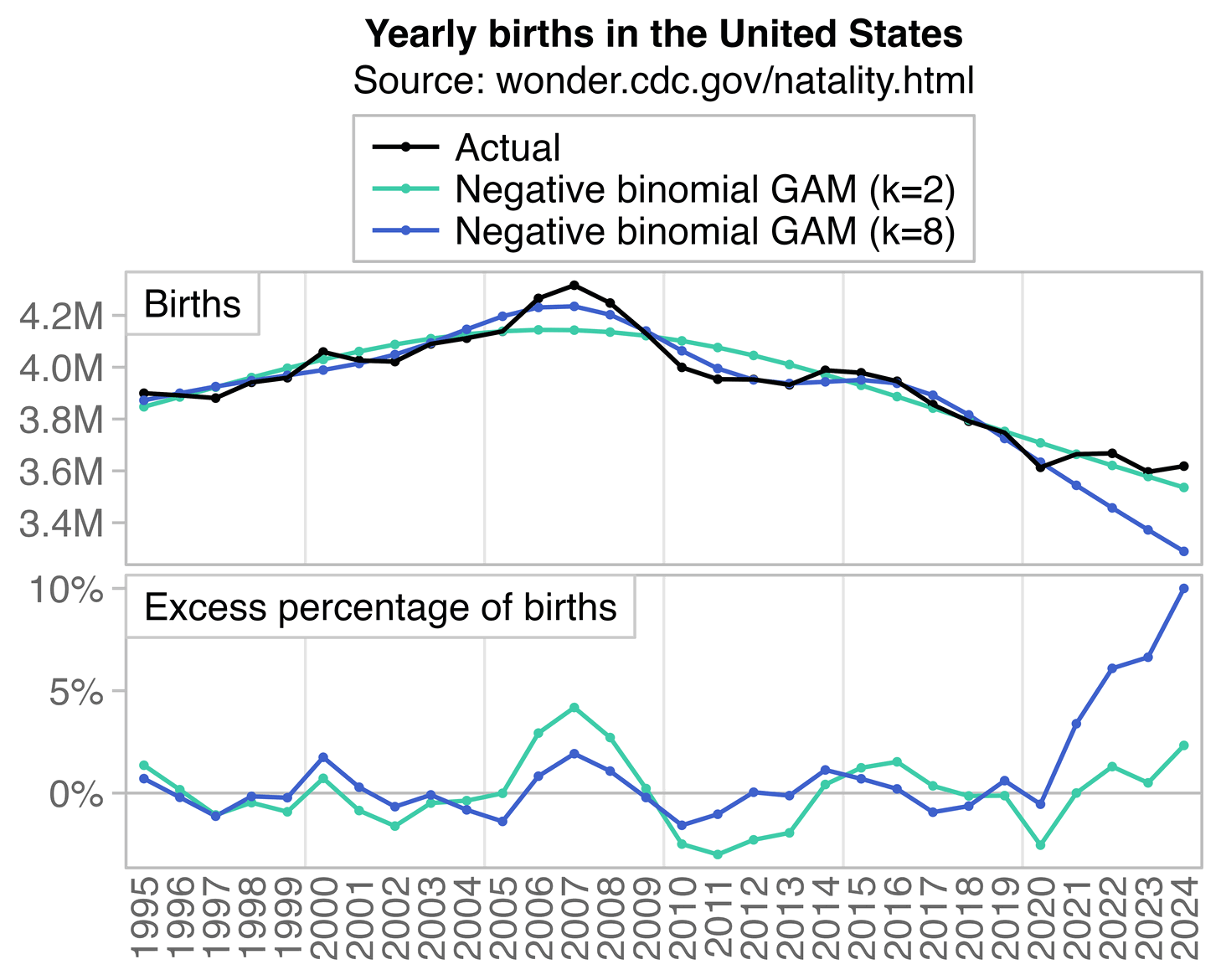
library(mgcv)
t=fread("https://sars2.net/f/wonderinfantsexborn.csv")
p1=t[,.(y=sum(born),z=1),.(x=year)]
p2=p1[,.(x,z=2,y=as.double(predict(gam(y~s(x,k=2),nb(),.SD[x<2020],method="REML"),.SD,type="response")))]
p3=p1[,.(x,z=3,y=as.double(predict(gam(y~s(x,k=8),nb(),.SD[x<2020],method="REML"),.SD,type="response")))]
p=rbind(p1,p2,p3)
p[,z:=factor(z,,c("Actual","Negative binomial GAM (k=2)","Negative binomial GAM (k=8)"))]
p=rbind(p[,facet:=1],p[,.(y=(y[1]/y-1)*100,z),x][z!=z[1]][,facet:=2])
p[,facet:=factor(facet,,c("Births","Excess percentage of births"))]
xstart=1994.5;xend=2024.5;xbreak=seq(1995,2024,1)
p=p[x>=xstart&x<=xend]
ylim=p[,{x=extendrange(y);.(min=x[1],max=x[2])},facet]
ggplot(p)+
facet_wrap(~facet,ncol=1,dir="v",scales="free_y")+
geom_vline(xintercept=seq(1999.5,xend,5),color="gray90",linewidth=.4)+
geom_rect(data=ylim,aes(ymin=min,ymax=max),xmin=xstart,xmax=xend,lineend="square",linejoin="mitre",fill=NA,color="gray75",linewidth=.4)+
geom_segment(data=ylim[2],aes(xstart,0),xend=xend,yend=0,color="gray75",linewidth=.4)+
geom_line(aes(x,y,color=z),linewidth=.6)+
geom_point(aes(x,y,color=z),stroke=0,size=1.3)+
geom_line(data=p[z==z[1]],aes(x,y),linewidth=.6)+
geom_label(data=ylim,aes(xstart,max,label=facet),vjust=1,hjust=0,label.r=unit(0,"pt"),label.padding=unit(5,"pt"),label.size=.4,size=3.87,fill="white",color="gray80")+
geom_label(data=ylim,aes(xstart,max,label=facet),vjust=1,hjust=0,label.r=unit(0,"pt"),label.padding=unit(5,"pt"),label.size=0,size=3.87,fill=NA)+
labs(title="Yearly births in the United States",subtitle="Source: wonder.cdc.gov/natality.html",x=NULL,y=NULL)+
scale_x_continuous(limits=c(xstart,xend),breaks=xbreak,labels=xbreak)+
scale_y_continuous(breaks=\(x)pretty(x,5),labels=\(x)if(max(x,na.rm=T)<1e3)paste0(x,"%")else sprintf("%.1fM",x/1e6))+
scale_color_manual(values=c("black",hsl(c(180,240)-15,70,80)))+
coord_cartesian(clip="off",expand=F)+
theme(axis.text=element_text(size=11,color="gray40",margin=margin(2,2,2,2)),
axis.text.x=element_text(angle=90,vjust=.5,hjust=1),
axis.ticks=element_line(linewidth=.4,color="gray75"),
axis.ticks.length.x=unit(0,"pt"),
axis.ticks.length.y=unit(4,"pt"),
legend.background=element_rect(color="gray75",linewidth=.4),
legend.box="vertical",
legend.box.margin=margin(,,3),
legend.box.spacing=unit(0,"pt"),
legend.direction="vertical",
legend.key=element_blank(),
legend.key.height=unit(12,"pt"),
legend.key.width=unit(24,"pt"),
legend.margin=margin(3,5,3,3),
legend.position="top",
legend.spacing.x=unit(2,"pt"),
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=11),
legend.title=element_blank(),
panel.background=element_blank(),
panel.grid=element_blank(),
panel.spacing=unit(3,"pt"),
plot.subtitle=element_text(size=11,hjust=.5,margin=margin(,,4)),
plot.title=element_text(size=11,hjust=.5,face="bold",margin=margin(,,3)),
strip.background=element_blank(),
strip.text=element_blank())
ggsave("1.png",width=4.86,height=3.9,dpi=300*4)
So if the high k-value is more accurate according to McCairn, then did vaccines cause a massive baby boom? Or is McCairn now in favor of using a low k-value instead, because he simply wants me to use whatever k-value supports his agenda?
Clare Craig made the plot below which shows a ratio between whites and blacks for infant deaths divided by mid-year resident population estimates. She found a sudden change in the slope of the ratio between 2019 and 2020: [https://drclarecraig.substack.com/p/battle-of-the-baselines]
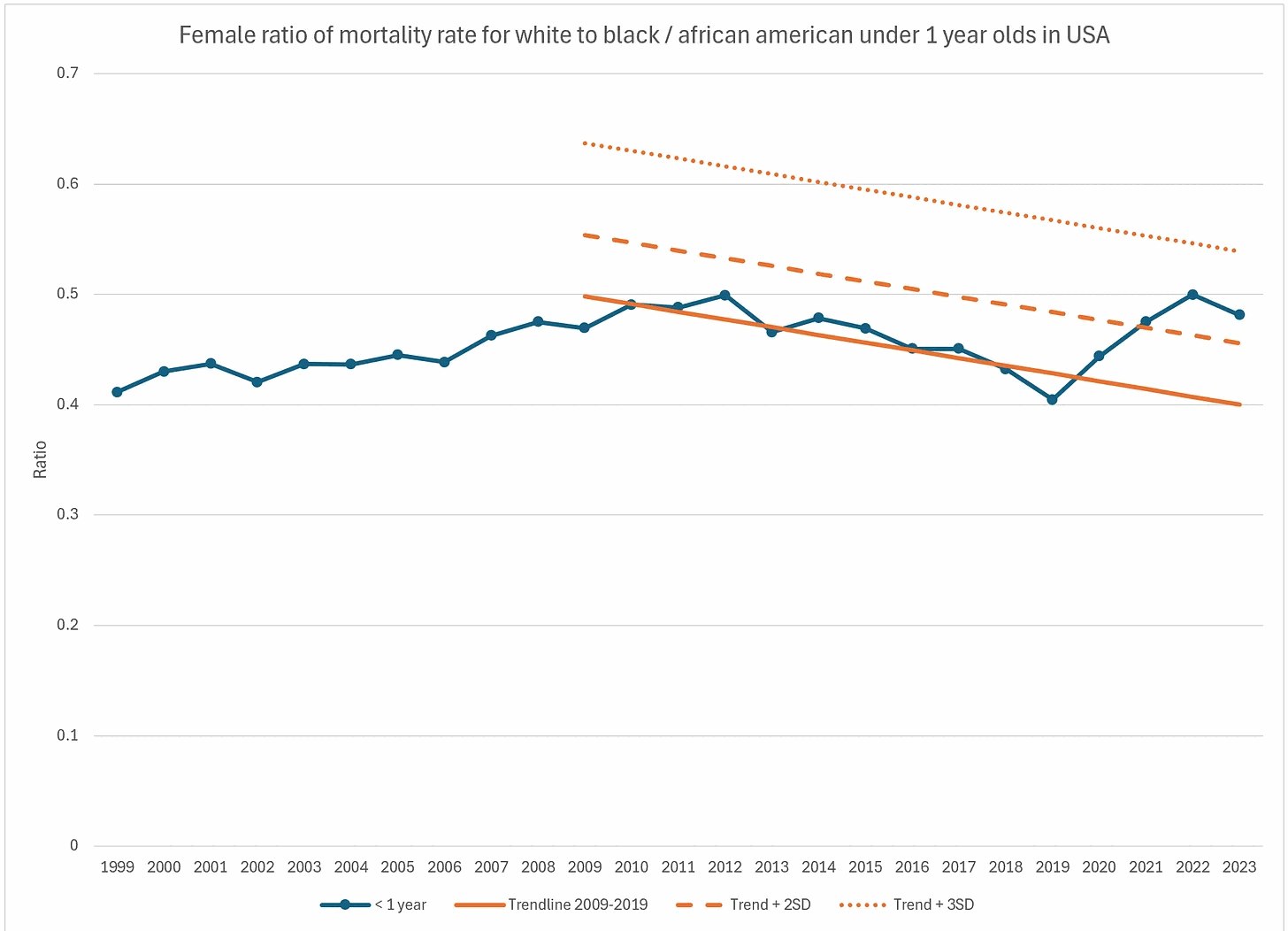
In the top left panel of the next plot, I used the population estimates returned by CDC WONDER like Clare Craig. CDC WONDER uses population estimates based on the 2010 census for 2011-2020 and population estimates based on the 2020 census for 2021-2024, but the 2010-based estimates are not adjusted to get rid of a sudden jump in population size after the switch of census base. CDC WONDER also uses vintage 2021 population estimates for 2021, even though the vintage 2024 estimates for 2021 are likely more accurate, and the Census Bureau recommends using the latest-vintage estimates for each year. In the top right panel I used the latest-vintage estimates for each year, and I generated DIY intercensal estimates for 2010-2019 to get rid of the jump after the census base, which got rid of the sudden change in the slope of the ratio between the late 2010s and early 2020s:
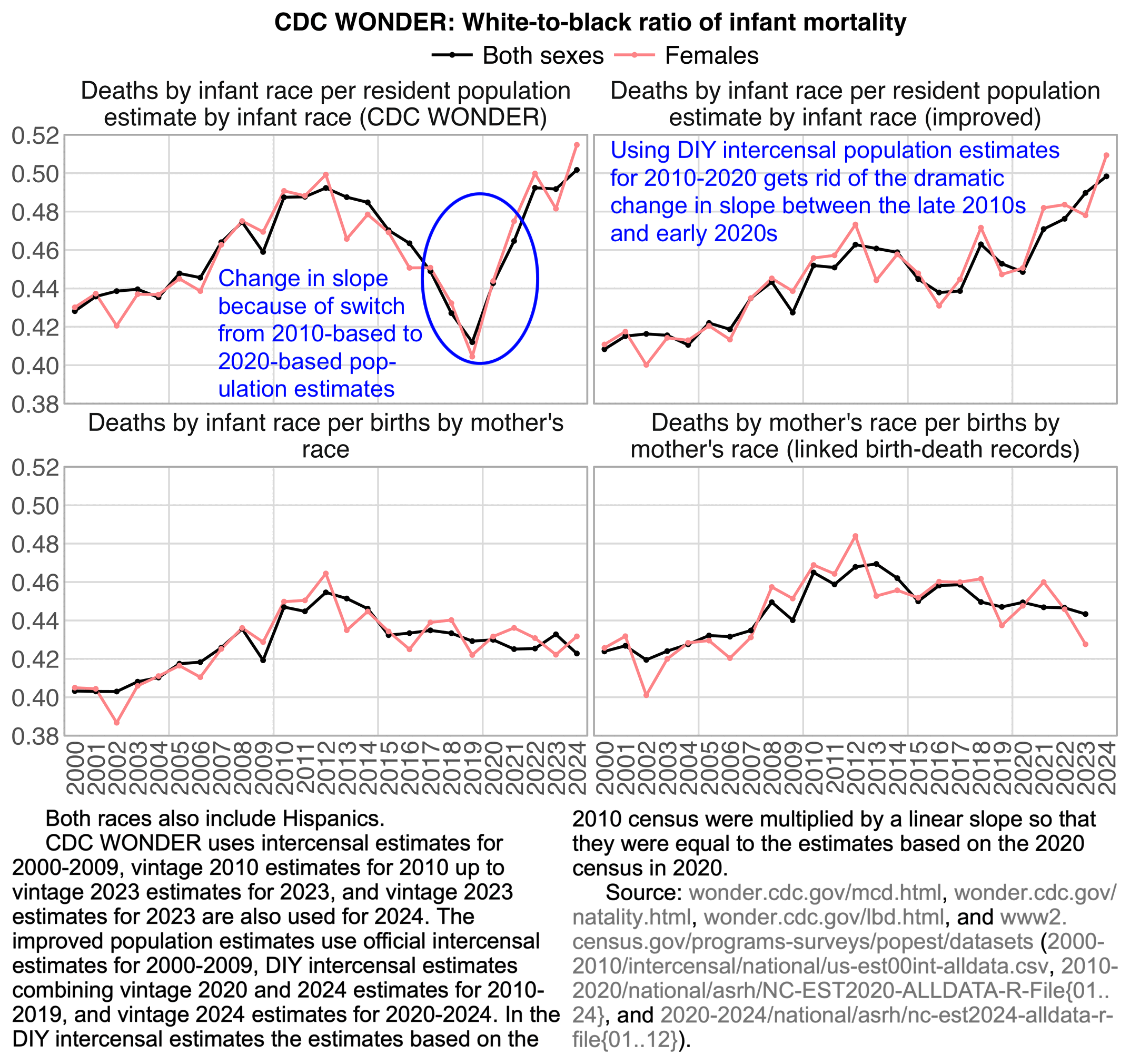
u="https://www2.census.gov/programs-surveys/popest/datasets/"
pop1=fread(paste0(u,"2000-2010/intercensal/national/us-est00int-alldata.csv"))
pop2=do.call(rbind,lapply(sprintf("%s2010-2020/national/asrh/NC-EST2020-ALLDATA-R-File%02d.csv",u,1:24),fread))
pop3=do.call(rbind,lapply(sprintf("%s2020-2024/national/asrh/nc-est2024-alldata-r-file%02d.csv",u,1:12),fread))
pop=rbind(pop1[YEAR<2010,1:10][,type:=1],pop2[,2:11][,type:=2],pop3[,2:11][,type:=3])
pop=pop[AGE==0&MONTH==7,.(pop=c(WA_MALE,WA_FEMALE,BA_MALE,BA_FEMALE),race=rep(c("White","Black"),each=.N*2),sex=rep(c("Male","Female"),each=.N),year=YEAR,type)]
pop=merge(pop,pop[year==2020,.(ratio=pop[type==3]/pop[type==2]),.(race,sex)])
pop=pop[!(type==2&year==2020)][,pop:=as.double(pop)]
pop[type==2,pop:={x=(year-2010)/10;pop*(x*ratio+1-x)}]
t=fread("https://sars2.net/f/wonderinfantyearracesex.csv")
t[race=="Black or African American",race:="Black"]
t=rbind(t,merge(t[type=="pop",.(year,sex,race,dead,type="newpop")],pop[,.(year,sex,race,pop)]))
t=rbind(cbind(t,z=1),t[sex=="Female"][,z:=2])
p=t[,.(sum(dead)/sum(pop)),.(race,year,z,type)]
p=p[,.(y=V1[race=="White"]/V1[race=="Black"]),.(z,facet=type,x=year)]
p[,z:=factor(z,,c("Both sexes","Females"))]
p[,facet:=factor(facet,c("pop","newpop","born","linked"),str_wrap(c("Deaths by infant race per resident population estimate by infant race (CDC WONDER)","Deaths by infant race per resident population estimate by infant race (improved)","Deaths by infant race per births by mother's race","Deaths by mother's race per births by mother's race (linked birth-death records)"),45))]
p=na.omit(p[x%in%2000:2024])
xmin=1999.5;xmax=2024.5;xbreak=2000:2024
ylim=cbind(facet=p[rowid(facet)==1,facet],p[,.(min=min(y),max=max(y))])
ybreak=pretty(p$y);ymin=min(ybreak);ymax=max(ybreak)
ggplot(p)+
facet_wrap(~facet,ncol=2,dir="h")+
geom_vline(xintercept=seq(xmin,xmax,5),color="gray90",linewidth=.4)+
geom_hline(yintercept=ybreak,color="gray90",linewidth=.4)+
annotate("rect",ymin=ymin,ymax=ymax,xmin=xmin,xmax=xmax,lineend="square",linejoin="mitre",fill=NA,color="gray72",linewidth=.4)+
geom_line(aes(x,y,color=z),linewidth=.6)+
geom_point(aes(x,y,color=z),stroke=0,size=1.2)+
labs(title="CDC WONDER: White-to-black ratio of infant mortality",x=NULL,y=NULL)+
scale_x_continuous(breaks=xbreak)+
scale_y_continuous(breaks=ybreak)+
scale_color_manual(values=c("black",hsv(0,.4,1)))+
coord_cartesian(clip="off",expand=F)+
theme(axis.text=element_text(size=11,color="gray40",margin=margin(2,2,2,2)),
axis.text.x=element_text(angle=90,vjust=.5,hjust=1),
axis.ticks.length=unit(0,"pt"),
legend.background=element_blank(),
legend.box.spacing=unit(0,"pt"),
legend.direction="horizontal",
legend.key=element_blank(),
legend.key.height=unit(13,"pt"),
legend.key.width=unit(23,"pt"),
legend.margin=margin(,,4),
legend.position="top",
legend.spacing.x=unit(2,"pt"),
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=11,vjust=.5),
legend.title=element_blank(),
panel.background=element_blank(),
panel.grid=element_blank(),
panel.spacing=unit(3,"pt"),
plot.title=element_text(size=11,hjust=.5,face="bold",margin=margin(,,2)),
strip.background=element_blank(),
strip.text=element_text(size=11,margin=margin(,,3)))
ggsave("1.png",width=7,height=5,dpi=300*4)
CDC WONDER's population estimates often have large jumps year-to-year, because CDC WONDER uses the earliest-vintage and not latest-vintage estimates, and it doesn't have intercensal estimates for 2010-2020. The next plot shows that in CDC WONDER's population estimates for age 0, blacks have a huge drop in population size between 2017 and 2018. And also between 2019 and 2020, the population size of whites goes down but the population size of blacks goes up, even though both races had a reduction in births between 2019 and 2020:
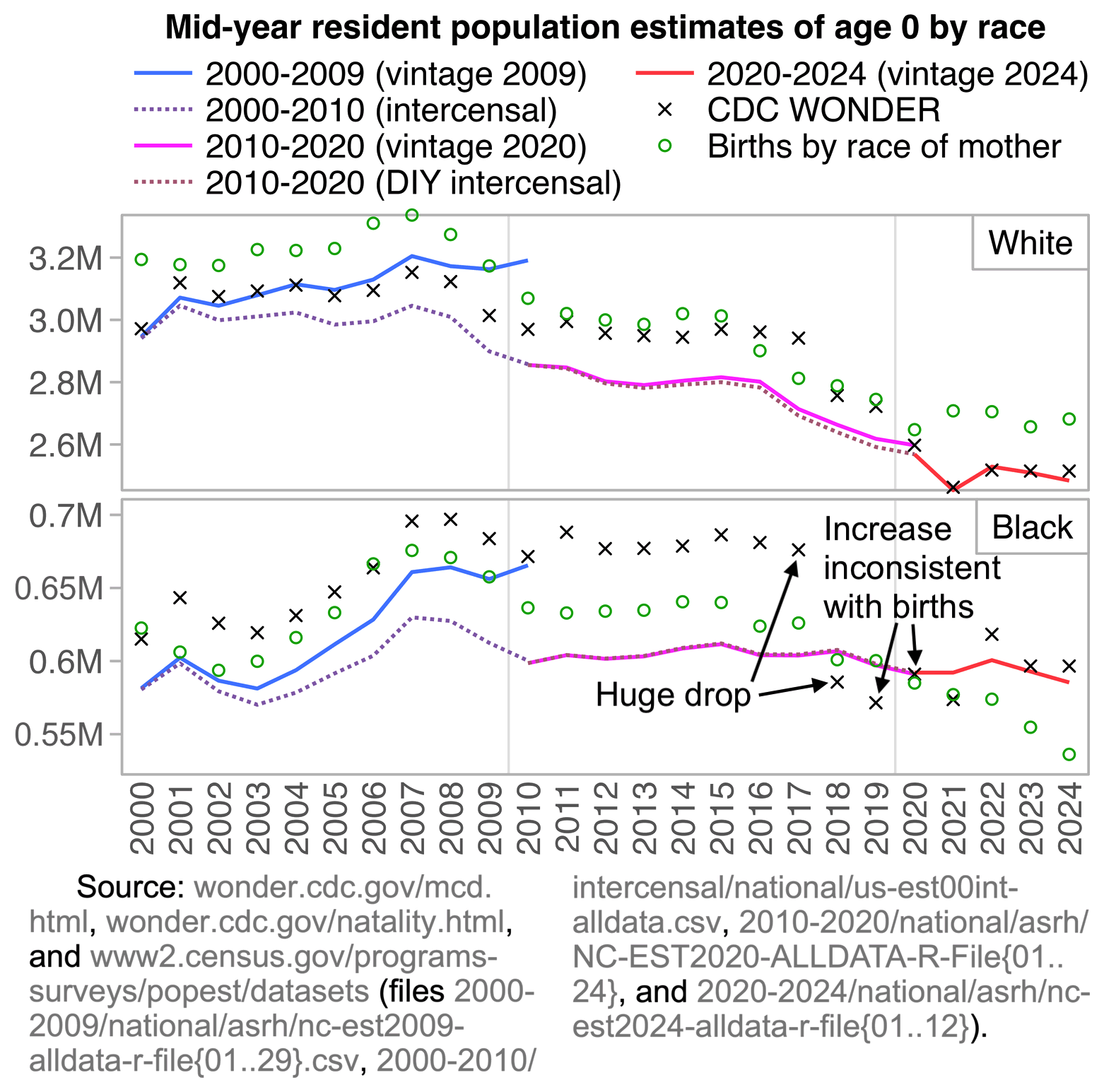
u="https://www2.census.gov/programs-surveys/popest/"
pop0=do.call(rbind,lapply(sprintf("%sdatasets/2000-2009/national/asrh/nc-est2009-alldata-r-file%02d.csv",u,1:29),fread))
pop1=fread(paste0(u,"datasets/2000-2010/intercensal/national/us-est00int-alldata.csv"))
pop2=do.call(rbind,lapply(sprintf("%sdatasets/2010-2020/national/asrh/NC-EST2020-ALLDATA-R-File%02d.csv",u,1:24),fread))
pop3=do.call(rbind,lapply(sprintf("%sdatasets/2020-2024/national/asrh/nc-est2024-alldata-r-file%02d.csv",u,1:12),fread))
pop=rbind(pop0[MONTH==7&AGE==0][-.N,2:11][,type:=0],pop1[,1:10][,type:=1],pop2[YEAR<2021,2:11][,type:=2],pop3[,2:11][,type:=4])
pop=pop[AGE==0&MONTH==7,.(pop=c(WA_MALE,WA_FEMALE,BA_MALE,BA_FEMALE),race=rep(c("White","Black"),each=.N*2),sex=rep(c("Male","Female"),each=.N),year=YEAR,type)]
pop=merge(pop,pop[year==2020,.(ratio=pop[type==4]/pop[type==2]),.(race,sex)])
pop[,pop:=as.double(pop)]
pop=rbind(pop,pop[type==2][,type:=3][,pop:={x=(year-2010)/10;pop*(x*ratio+1-x)}])
p=pop[,.(y=sum(pop)),.(x=year,z=type,facet=race)]
t=fread("https://sars2.net/f/wonderinfantyearracesex.csv")
t[race=="Black or African American",race:="Black"]
p=rbind(p,t[type=="pop",.(y=sum(pop),z=5),.(x=year,facet=race)])
p=rbind(p,t[type=="born",.(y=sum(pop),z=6),.(x=year,facet=race)])
race=c("White","Black")
p=p[facet%in%race][,facet:=factor(facet,race)]
p[,z:=factor(z,,c("2000-2009 (vintage 2009)","2000-2010 (intercensal)","2010-2020 (vintage 2020)","2010-2020 (DIY intercensal)","2020-2024 (vintage 2024)","CDC WONDER","Births by race of mother"))]
p=na.omit(p[x%in%2000:2024])
xmin=1999.5;xmax=2024.5;xbreak=2000:2024
ylim=p[,.(min=min(y),max=max(y)),facet]
rat=ylim[,max(max/min)]
ylim=ylim[,{x=(rat*min-max)/(1+rat);.(min=min-x,max=max+x,facet)}]
ggplot(p)+
facet_wrap(~facet,ncol=1,scales="free_y")+
geom_vline(xintercept=seq(xmin,xmax,10),color="gray90",linewidth=.4)+
geom_rect(data=ylim,aes(ymin=min,ymax=max),xmin=xmin,xmax=xmax,lineend="square",linejoin="mitre",fill=NA,color="gray75",linewidth=.4)+
geom_line(aes(x,y,color=z,linewidth=z,linetype=z))+
geom_point(aes(x,y,color=z,shape=z,size=z),stroke=.5)+
geom_label(data=ylim,aes(xmax,max,label=facet),hjust=1,vjust=1,fill="white",label.r=unit(0,"pt"),label.padding=unit(5,"pt"),label.size=.4,color="gray75",size=3.87)+
geom_label(data=ylim,aes(xmax,max,label=facet),hjust=1,vjust=1,fill=NA,label.r=unit(0,"pt"),label.padding=unit(5,"pt"),size=3.87,label.size=0)+
labs(title="Mid-year resident population estimates of age 0 by race",x=NULL,y=NULL)+
scale_x_continuous(breaks=xbreak)+
scale_y_continuous(breaks=pretty,labels=\(x)sprintf("%.1fM",x/1e6))+
scale_color_manual(values=c(hsv(c(22,27,30,34,0)/36,c(.7,.4,.7,.4,.7),c(1,.7,1,.7,1)),"black","#00aa00"))+
scale_shape_manual(values=c(NA,NA,NA,NA,NA,4,1))+
scale_linetype_manual(values=c("solid","11")[c(1,2,1,2,1,1,1)])+
scale_alpha_manual(values=c(0,0,0,0,0,1,1))+
scale_linewidth_manual(values=c(.6,.6,.6,.6,.6,0,0))+
scale_size_manual(values=rep(c(1.1,1.4),c(5,2)))+
guides(color=guide_legend(ncol=2,byrow=F))+
coord_cartesian(clip="off",expand=F)+
theme(axis.text=element_text(size=11,color="gray40",margin=margin(2,2,2,2)),
axis.text.x=element_text(angle=90,vjust=.5,hjust=1),
axis.ticks=element_line(linewidth=.4,color="gray75"),
axis.ticks.length.x=unit(0,"pt"),
axis.ticks.length.y=unit(4,"pt"),
legend.background=element_blank(),
legend.box.spacing=unit(0,"pt"),
legend.direction="horizontal",
legend.key=element_blank(),
legend.key.height=unit(12,"pt"),
legend.key.width=unit(24,"pt"),
legend.margin=margin(,,5),
legend.position="top",
legend.spacing.x=unit(2,"pt"),
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=11,vjust=.5),
legend.title=element_blank(),
panel.background=element_blank(),
panel.grid=element_blank(),
panel.spacing=unit(3,"pt"),
plot.title=element_text(size=11,hjust=.5,face="bold",margin=margin(,,3)),
strip.background=element_blank(),
strip.text=element_blank())
ggsave("1.png",width=5,height=4,dpi=300*4)
In the plot above the black births are lower than the black population estimates since 2020, but it might be because I looked at births by maternal race, and some infants with a black father but a non-black mother might be classified as black in the population estimates.
In the next plot I retrieved deaths from CDC WONDER by infant cause of death groups ("MCD - ICD-10 130 Cause List (Infants)"). Deaths involving viral diseases or respiratory conditions seem to have increased between 2021 and 2022, which might partially be because respiratory viruses entered back in circulation after lockdown measures were lifted and people stopped wearing masks:
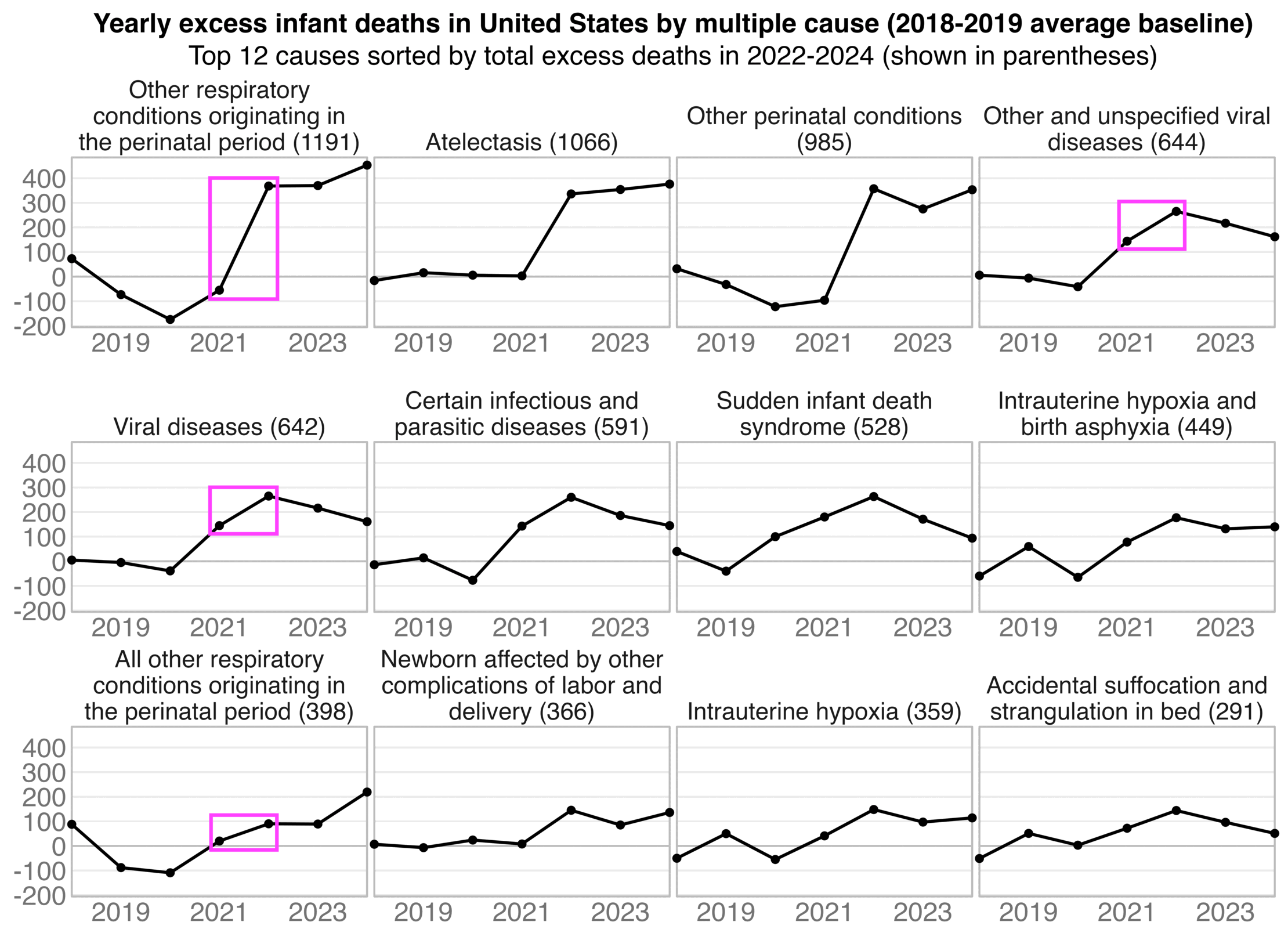
t=fread("https://sars2.net/f/wonderinfantcausemcd.csv")[year<2025] # MCD
# t=fread("https://sars2.net/f/wonderinfantcausesex.csv")[year<2025] # UCD
p=t[,.(y=sum(dead)),.(x=year,facet=sub(" \\([^(]*\\)$","",cause))]
p=p[facet%in%p[,.N,facet][N==max(N),facet]]
p[,y:=y-mean(y[x<2020]),facet]
p[,facet:=sub(", not elsewhere classified","",facet)]
sum=p[x%in%2022:2024,.(y=sum(y)),facet][order(-y)][1:12]
p=p[facet%in%sum$facet][,facet:=factor(facet,sum$facet,str_wrap(sum[,paste0(facet," (",y,")")],27))]
xstart=2018;xend=2024;xbreak=seq(2019,2024,2)
ylim=p[,{x=extendrange(y);.(min=x[1],max=x[2])}]
ggplot(p)+
facet_wrap(~facet,ncol=4,dir="h",scales="free_x")+
geom_hline(yintercept=0,color="gray75",linewidth=.4)+
geom_rect(data=ylim,aes(ymin=min,ymax=max),xmin=xstart,xmax=xend,lineend="square",linejoin="mitre",fill=NA,color="gray75",linewidth=.4)+
geom_line(aes(x,y),linewidth=.6)+
geom_point(aes(x,y),size=1.1)+
labs(x=NULL,y=NULL,title="Yearly excess infant deaths in United States by multiple cause (2018-2019 average baseline)",subtitle="Top 12 causes sorted by total excess deaths in 2022-2024 (shown in parentheses)")+
scale_x_continuous(limits=c(xstart,xend),breaks=xbreak)+
scale_y_continuous(breaks=pretty)+
coord_cartesian(clip="off",expand=F)+
theme(axis.text=element_text(size=11,color="gray45"),
axis.ticks.length=unit(0,"pt"),
legend.background=element_blank(),
legend.box.spacing=unit(0,"pt"),
legend.key=element_blank(),
legend.key.height=unit(12,"pt"),
legend.key.width=unit(24,"pt"),
legend.margin=margin(,,4),
legend.position="top",
legend.spacing.x=unit(2,"pt"),
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=11),
legend.title=element_blank(),
panel.background=element_blank(),
panel.grid=element_blank(),
panel.grid.major.y=element_line(linewidth=.4,color="gray92"),
panel.spacing=unit(3,"pt"),
plot.subtitle=element_text(size=11,hjust=.5,margin=margin(,,4)),
plot.title=element_text(size=11,hjust=.5,face=2,margin=margin(,,3)),
strip.background=element_blank(),
strip.text=element_text(hjust=.5,size=10,margin=margin(-2,,3),vjust=0))
ggsave("1.png",width=7.4,height=5.4,dpi=300*4)
Infants also had more COVID deaths in 2022 than 2021:
| Year | Deaths at age 0 with UCD U07.1 | Deaths at age 0 with MCD U07.1 |
|---|---|---|
| 2020 | 35 | 52 |
| 2021 | 91 | 167 |
| 2022 | 141 | 248 |
| 2023 | 59 | 99 |
| 2024 | 53 | 101 |
ES posted this response to people like me who requested him to document his methodology: [https://x.com/EthicalSkeptic/status/1962179284451156283]
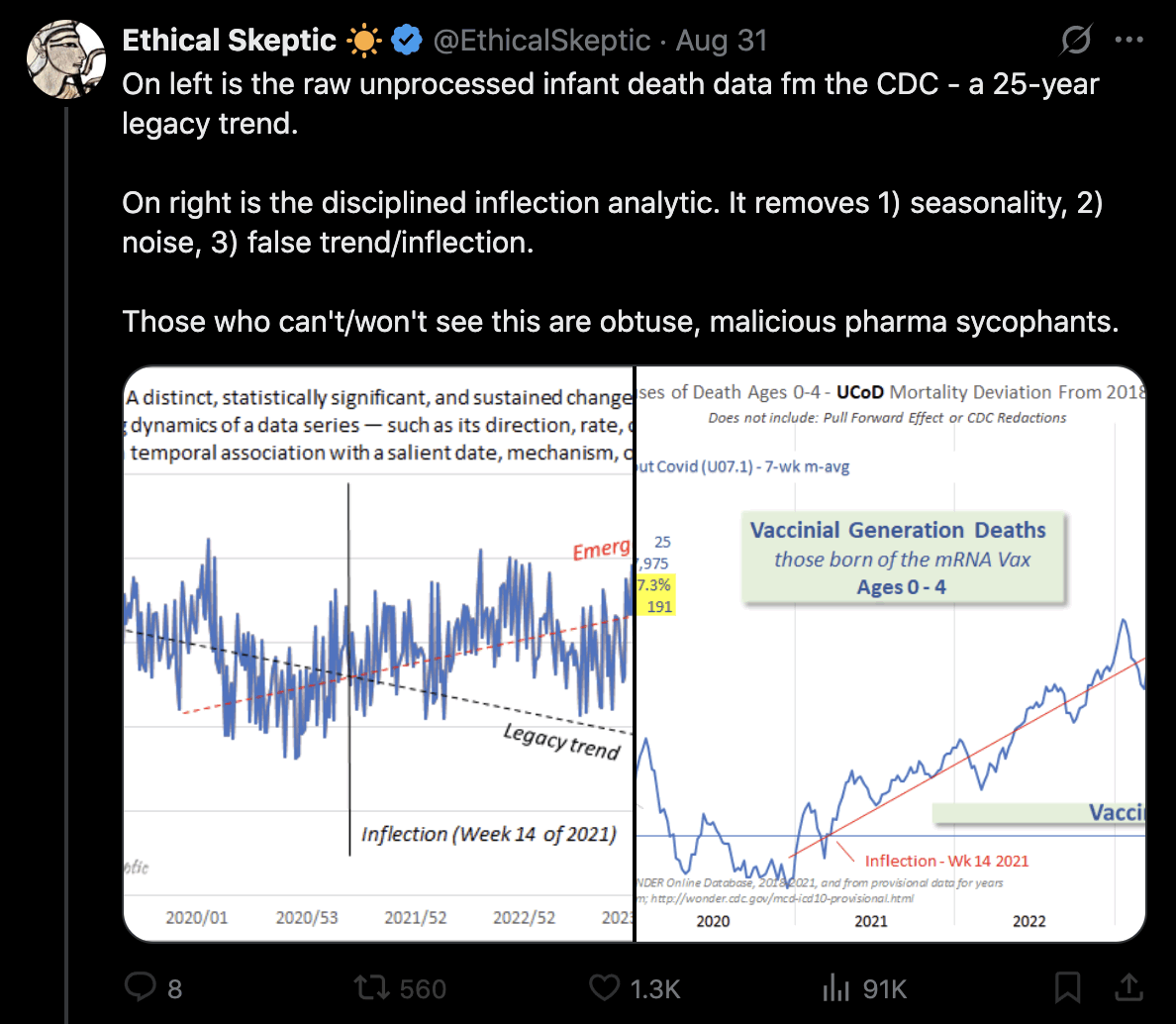
However if he doesn't want to publish his Excel spreadsheets, he could simply describe his methodology precisely enough that other people can reproduce his weekly number of excess deaths exactly. Or he could write a script in some programming language, so that if it's given the weekly number of deaths as an input, it produces the weekly baseline number of deaths as the output. It probably wouldn't require 150 modules to accomplish.
I have posted over a thousand R scripts on my website, and I have done a lot of extra work to write my R scripts in a format where other people can run them by copying and pasting my code. If Ethical Skeptic's Excel-based workflow is not as easy to reproduce or to share with people, then it's one way in which his analysis is inferior to mine.
Someone offered to reproduce Ethical Skeptic's plot in Python or R and make his code open source, but ES refused to help him reproduce the plot: [https://x.com/DrNo_Reformed/status/1962214582107164675]
I bet if Ethical Skeptic would write a scientific paper, the methods section would say: "The methodology is immaterial. You will find the way once you understand the principles of epoché and ponerology. I have over 150 modules, so my methods are impossible to document."
Apparently even ChatGPT recognized that Ethical Skeptic's methodology is too sophisticated to be shared at GitHub: [https://x.com/EthicalSkeptic/status/1971245590877987321]
ES claimed that "Our children are now dying at rates we've never seen before": [https://x.com/EthicalSkeptic/status/1963365670613127206]
In the following code I took the yearly number of deaths by single year of age from CDC WONDER, and I used the Census Bureau's mid-year resident population estimates with DIY intercensal adjustment for 2011-2019. [ethical.html#Files_for_US_population_estimates_by_single_year_of_age] The all-cause ASMR of ages 0-4 was lower in 2024 than any earlier year apart from 2019 and 2020:
t=fread("https://sars2.net/f/uspopdead.csv")[year%in%2010:2024]
# use total person-years in 2010-2024 as the standard population
t[,std:=(tapply(pop,age,sum)/sum(pop))[age+1]]
# ASMR per 100,000 people aged 0-4
t[age<5,.(asmr=round(sum(dead/pop*std*1e5),2)),year]
# year asmr
# 2010 8.59
# 2011 8.38
# 2012 8.34
# 2013 8.24
# 2014 8.07
# 2015 8.13
# 2016 8.11
# 2017 7.96
# 2018 7.79
# 2019 7.70
# 2020 7.30
# 2021 7.75
# 2022 7.90
# 2023 7.78
# 2024 7.74
The next plot shows that there were spikes in the rate of stillbirths per live births during COVID waves. There was a particularly high peak during the Delta wave in September 2021, when there was also a high number of deaths in women of childbearing age. But after the first Omicron wave in early 2022, the rate of stillbirths per live births roughly fell on the pre-COVID trend: [https://wonder.cdc.gov/fetal.html]
t=fread("https://sars2.net/f/wondernatality.csv")
t=merge(t,fread("https://sars2.net/f/wonderstillbirths.csv"))
a=t[,.(mdead=mean(dead),mborn=mean(born)),year]
a=merge(a,t)[,.(mdead=mean(dead-mdead),mborn=mean(born-mborn)),month]
t=merge(a,t)[,.(year,month,dead=dead-mdead,born=born-mborn)]
t[,x:=as.Date(paste0(year,"-",month,"-16"))]
p=t[,.(x,y=c(dead,born,dead/born*1e3),z=rep(1:3,each=.N))]
p[,z:=factor(z,,c("Stillbirths","Live births","Stillbirths per 1,000 live births"))]
xstart=as.Date("2005-1-1");xend=as.Date("2024-1-1");xbreak=seq(xstart+182,xend,"year")
ylim=p[,{x=extendrange(y);.(min=x[1],max=x[2])},z]
rat=ylim[,max(max/min)]
ylim=ylim[,{x=(rat*min-max)/(1+rat);.(min=min-x,max=max+x,z)}]
ggplot(p)+
facet_wrap(~z,ncol=1,dir="v",scales="free_y")+
geom_vline(xintercept=seq(xstart,xend,"year"),color="gray90",linewidth=.4)+
geom_rect(data=ylim,aes(ymin=min,ymax=max),xmin=xstart,xmax=xend,lineend="square",linejoin="mitre",fill=NA,color="gray75",linewidth=.4)+
geom_line(aes(x,y),linewidth=.6)+
geom_label(data=ylim,aes(xend,max,label=z),vjust=1,hjust=1,label.r=unit(0,"pt"),label.padding=unit(4,"pt"),label.size=.4,size=3.87,fill="white",color="gray80")+
geom_label(data=ylim,aes(xend,max,label=z),vjust=1,hjust=1,label.r=unit(0,"pt"),label.padding=unit(4,"pt"),label.size=0,size=3.87,fill=NA)+
labs(title="Stillbirths at CDC WONDER (seasonal component removed)",x=NULL,y=NULL)+
scale_x_continuous(limits=c(xstart,xend),breaks=xbreak,labels=year(xbreak))+
scale_y_continuous(breaks=\(x)pretty(x,4),labels=\(x)ifelse(x>=1e3,paste0(x/1e3,"k"),x))+
coord_cartesian(clip="off",expand=F)+
theme(axis.text=element_text(size=11,color="gray40",margin=margin(2,2,2,2)),
axis.text.x=element_text(angle=90,vjust=.5,hjust=1),
axis.ticks=element_line(linewidth=.4,color="gray75"),
axis.ticks.length.x=unit(0,"pt"),
axis.ticks.length.y=unit(4,"pt"),
panel.background=element_blank(),
panel.grid=element_blank(),
panel.spacing=unit(3,"pt"),
plot.title=element_text(size=11,hjust=.5,face="bold",margin=margin(1,,4)),
strip.background=element_blank(),
strip.text=element_blank())
ggsave("1.png",width=5,height=3.6,dpi=300*4)
In the plot above I tried using a new method to remove a seasonal component from monthly data, where I don't have to take a moving average so I can preserve the original monthly temporal resolution, and I don't have to plot excess deaths relative to a seasonality-adjusted baseline. I don't know why I hadn't thought of the method before:
# demonstrate method with minimal data that includes only January and February t=CJ(year=2010:2011,month=1:2) t$born=c(323249,301994,320477,297961) # get yearly average births t=merge(t,t[,.(yearlyave=mean(born)),year]) # get average difference between actual births and yearly average for each month number t[,monthlyave:=tapply(born-yearlyave,month,mean)[month]] # subtract seasonal component round(t[,nonseasonal:=born-monthlyave]) # year month born yearlyave monthlyave nonseasonal # 2010 1 323249 312622 10943 312306 # 2010 2 301994 312622 -10943 312937 # 2011 1 320477 309219 10943 309534 # 2011 2 297961 309219 -10943 308904
ES linked to this image as an explanation of how he calculated his baseline: [https://theethicalskeptic.com/2025/08/19/houston-we-have-another-problem/]

He seems to have fitted his baseline against data starting from 2018 and not 2014, and he seems to have excluded certain weeks from his baseline fitting period, but I don't know if there's other ways how his methodology was different from the image above.
I attempted to replicate his methodology in the following code:
# get weekly deaths in ages 0-4 with the underlying cause A-R or 999--999
t=fread("https://sars2.net/f/wondervaccinial0to4.csv")
# convert deaths to a 6-week moving average
# the window extends up to 3 weeks backwards and up to 2 weeks forwards
ma=\(x,b=1,f=b)rowMeans(embed(c(rep(NA,b),x,rep(NA,f)),f+b+1),na.rm=T)
t[,dead:=ma(dead,3,2)]
# calculate the baseline using weeks of the MMWR years 2018 and 2019
# I excluded weeks 51 and 52 of 2019, but I don't know what weeks were actually excluded by ES
base=t[year<2020&!(year==2019&week>50)]
# get average deaths for each week number during the baseline period
# I treated week 53 as an average of weeks 52 and 1, but I don't know how ES treated week 53
weekly=base[,tapply(dead,week,mean)]
weekly[53]=mean(weekly[c(1,52)])
# get change in weekly average number of deaths between 2018 and 2019 (about -12.3)
# this is similar to the SLOPE function in Excel which was probably used by ES
slope=coef(lm(dead~year,base[year<2020]))[2]
# get annual growth rate relative to average weekly deaths in 2018-2019 (about -0.0295 or -3%)
growthrate=slope/base[year<2020,mean(dead)]
# get final baseline
t[,baseline:=weekly[week]*(1+growthrate)^(year-2018.5)]
If you do a linear regression of weekly deaths in 2018-2019 against a year variable, where there's 52 weeks where the value of the year variable is 2018 and 52 weeks where the value of the year variable is 2019, it's equivalent to doing a linear regression of yearly averages of weekly deaths, which is equivalent to simply subtracting the average deaths in 2018 from the average deaths in 2019:
t=fread("https://sars2.net/f/wondervaccinial0to4.csv")
# linear regression of weekly deaths against a year variable
coef(lm(dead~year,t[year<2020]))[2]
# -12.42308
# linear regression of yearly average deaths against a year variable
t[,.(dead=mean(dead)),year][year<2020,coef(lm(dead~year))[2]]
# -12.42308
# average deaths in 2019 minus average deaths in 2018
t[,.(dead=mean(dead)),year][,dead[year==2019]-dead[year==2018]]
# -12.42308
At first I wasn't sure how ES converted the slope of the linear regression to an annual growth percent, but then I noticed that in another image where he described his methodology, he wrote that he first determined "the first derivative dx/dy slope of 2014 - 2019 records for each ICD-10 code", and then he factored the "slope as a percentage of the average 2014 - 2019 week": [https://theethicalskeptic.com/wp-content/uploads/2025/09/ICD-10-Code-Excess-Death-Derivation-small.jpg]

So I think I now managed to otherwise reproduce the methodology described in his images, except I'm not sure if he only excluded weeks 51 and 52 of 2019 from the baseline fitting period, or if he also excluded other weeks. But I was still not successful in reproducing Ethical Skeptic's baseline, because I only got about 27% excess deaths on weeks 14-20 of 2025, and not 77%:
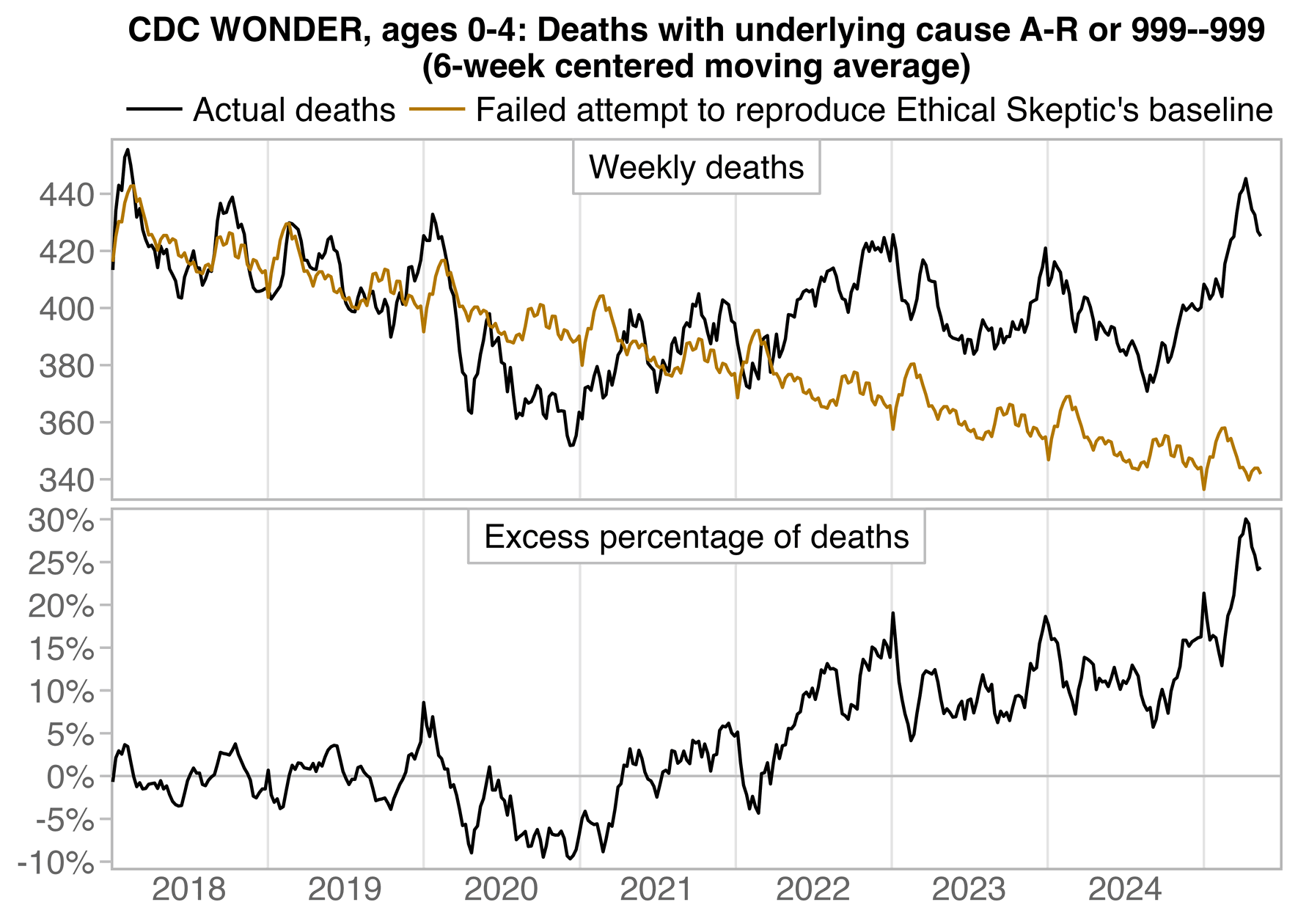
t=fread("https://sars2.net/f/wondervaccinial0to4.csv")
ma=\(x,b=1,f=b){x[]=rowMeans(embed(c(rep(NA,b),x,rep(NA,f)),f+b+1),na.rm=T);x}
t[,dead:=ma(dead,3,2)]
base=t[year<2020&!(year==2019&week>50)]
slope=coef(lm(dead~year,base[year<2020]))["year"]
growthrate=slope/base[year<2020,mean(dead)]
weekly=base[,tapply(dead,week,mean)]
weekly[53]=mean(weekly[c(1,52)])
t[,baseline:=weekly[week]*(1+growthrate)^(year-2018.5)]
t=t[!(year==2025&week>20)]
t[,x:=MMWRweek::MMWRweek2Date(year,week,4)]
p=t[,.(x,y=c(dead,baseline,(dead/baseline-1)*100),z=rep(c(1,2,1),each=.N),facet=rep(1:2,.N*2:1))]
p[,z:=factor(z,,c("Actual deaths","Failed attempt to reproduce Ethical Skeptic's baseline"))]
p[,facet:=factor(facet,,c("Weekly deaths","Excess percentage of deaths"))]
xstart=as.Date("2018-1-1");xend=as.Date("2025-7-1")
xbreak=seq(xstart,as.Date("2025-1-1"),"6 month");xlab=ifelse(month(xbreak)==7,year(xbreak),"")
ylim=p[,{x=extendrange(y,,.03);.(ymin=x[1],ymax=x[2])},facet]
ggplot(p)+
facet_wrap(~facet,ncol=1,scales="free_y")+
geom_vline(xintercept=seq(xstart,xend,"year"),color="gray90",linewidth=.4)+
geom_segment(data=p[.N],x=xstart,xend=xend,y=0,yend=0,linewidth=.4,color="gray75")+
geom_rect(data=ylim,aes(ymin=ymin,ymax=ymax),xmin=xstart,xmax=xend,lineend="square",linejoin="mitre",fill=NA,color="gray72",linewidth=.4)+
geom_line(aes(x,y,color=z),linewidth=.5)+
geom_label(data=ylim,aes(mean(c(xstart,xend)),ymax,label=facet),vjust=1,label.r=unit(0,"pt"),label.padding=unit(5,"pt"),label.size=.4,color="gray75",size=3.8)+
geom_label(data=ylim,aes(mean(c(xstart,xend)),ymax,label=facet),vjust=1,label.r=unit(0,"pt"),label.padding=unit(5,"pt"),label.size=0,size=3.8,fill=NA)+
labs(x=NULL,y=NULL,title="CDC WONDER, ages 0-4: Deaths with underlying cause A-R or 999--999\n(6-week centered moving average)")+
scale_x_date(limits=c(xstart,xend),breaks=xbreak,labels=xlab)+
scale_y_continuous(breaks=\(x)pretty(x,7),labels=\(x)if(min(x,na.rm=T)<0)paste0(x,"%")else x)+
scale_color_manual(values=c("black",hsv(4/36,1,.7)))+
coord_cartesian(clip="off",expand=F)+
theme(axis.text=element_text(size=11,color="gray40"),
axis.text.y=element_text(margin=margin(,1.5)),
axis.ticks=element_line(linewidth=.4,color="gray75"),
axis.ticks.length.x=unit(0,"pt"),
axis.ticks.length.y=unit(4,"pt"),
legend.background=element_blank(),
legend.box.spacing=unit(0,"pt"),
legend.direction="horizontal",
legend.key=element_blank(),
legend.key.height=unit(12,"pt"),
legend.key.spacing.x=unit(2,"pt"),
legend.key.width=unit(23,"pt"),
legend.margin=margin(-2,,4),
legend.position="top",
legend.spacing.x=unit(1,"pt"),
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=11,vjust=.5,margin=margin(,,,1)),
legend.title=element_blank(),
panel.background=element_blank(),
panel.spacing=unit(3,"pt"),
plot.subtitle=element_text(size=11,hjust=.5,margin=margin(,,4)),
plot.title=element_text(size=11,face=2,hjust=.5,margin=margin(,,4)),
strip.background=element_blank(),
strip.text=element_blank())
ggsave("1.png",width=5.9,height=4.2,dpi=300*4)
And anyway, there's no way you can get a baseline with Ethical
Skeptic's formula that decreases more between 2019 and 2020 than between
2018 and 2019, like the baseline in his plot for excess deaths in ages
0-4. If you give a negative growth rate to his formula
(1+growthrate)^(year-2018.5), you always get a concave
curve where the absolute year-to-year decrease gets smaller over
time:
d=CJ(year=2018:2025) growthrate=-.03 # 3% annual decrease d[,multiplier:=(1+growthrate)^(year-2018.5)] d[,diff:=c(NA,diff(multiplier))] round(d,4) # year multiplier diff # 2018 1.0153 NA # 2019 0.9849 -0.0305 # decrease between 2019 and 2020 is smaller # 2020 0.9553 -0.0295 # than decrease between 2018 and 2019 # 2021 0.9267 -0.0287 # 2022 0.8989 -0.0278 # 2023 0.8719 -0.0270 # 2024 0.8458 -0.0262 # 2025 0.8204 -0.0254
So that's yet another way I can tell that ES falsified his baseline, and he didn't actually employ the methodology he described in his image. (Or it's possible that he initially employed the methodology he described, but then he applied an additional downward adjustment to the baseline that he didn't document anywhere. But in any case he has not documented the complete methodology he used to derive the baseline.)
Around September 8th 2025 UTC, Ethical Skeptic edited his blog post to replace the plot with 77.3% excess deaths with a new plot that only had about 53.5% excess deaths on weeks 14-20 of 2025. [https://theethicalskeptic.com/2025/08/19/houston-we-have-another-problem/]
His caption for the new version of the plot said "chart revised 8 Sep 2025 to reflect greater conservancy and clean match to raw data in Chart 5 below". Chart 5 was the plot with the legacy trend and emerging trend, where the "legacy trend" appeared to approximate the trend in 2018-2020, even though the plot for excess deaths was meant to have a 2018-2019 baseline.
ES he didn't explain how he changed his methodology between the two versions of his plot. The number of deaths in 2018 and 2019 did not change between the two versions, so how was it possible for the slope of his baseline to change? If he would've actually calculated the baseline using the methodology described in his methodology image, I think the only variable he would've been able to change was the set of weeks in 2018 and 2019 that he excluded from the baseline fitting period. (But in reality I have shown that he falsified his baseline, because with the methodology described in his image, it wouldn't even be possible for the baseline to decrease more between 2019 and 2020 than between 2018 and 2019.)
But anyway, the new and old versions appear identical or close to identical in 2019. In general the shape of the lines within each year looks identical or close to identical, apart from the weeks where the window of the moving average includes weeks from the neighboring year. But ES seems to have simply altered the height of his baseline within each year as a whole:
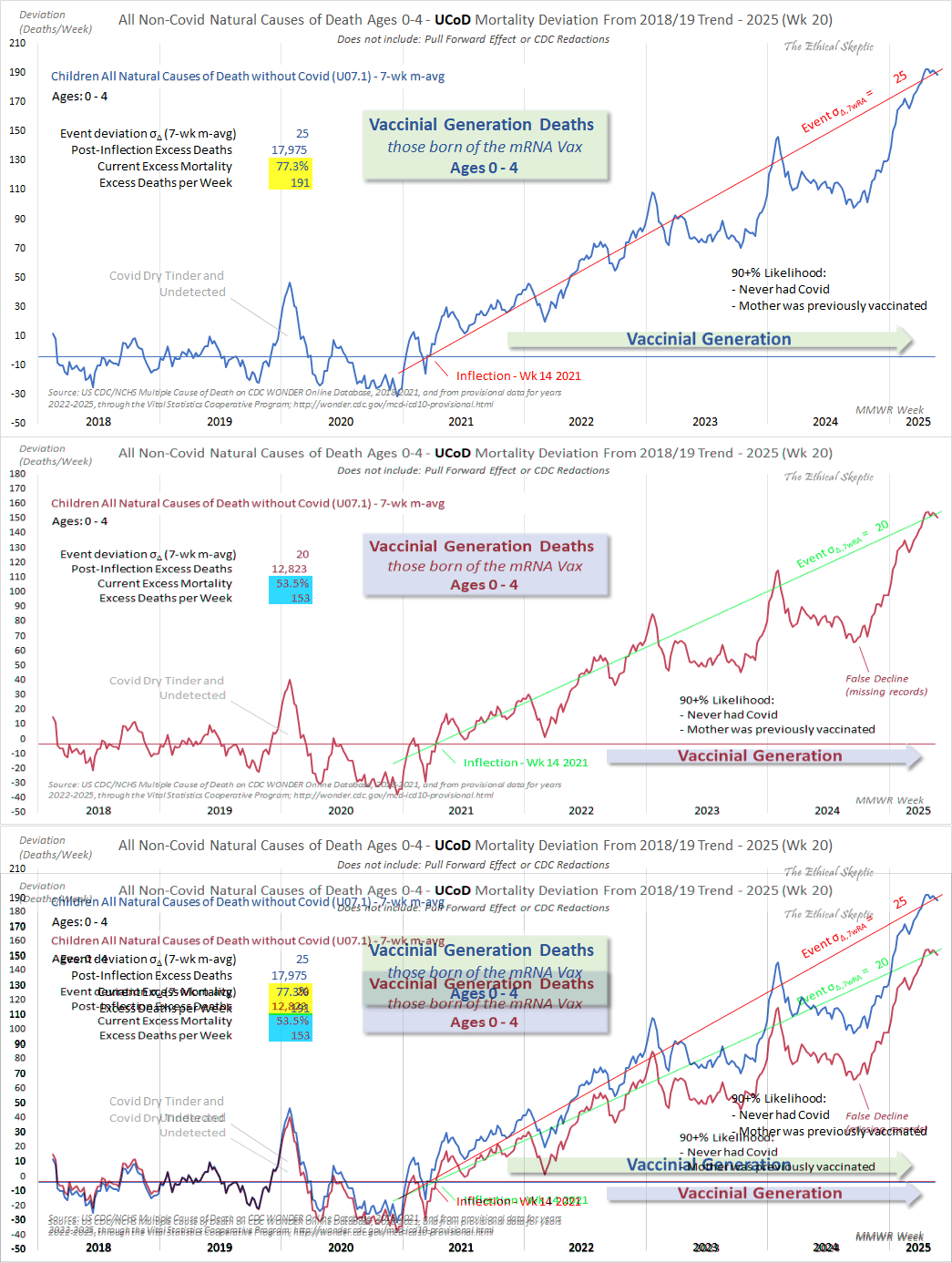
ES edited his blog post to add this screenshot of his spreadsheet: [https://theethicalskeptic.com/2025/08/19/houston-we-have-another-problem/]

His spreadsheet is missing the most crucial piece of information, which is how he calculated the baseline. I think ES knows himself that he falsified his baseline, so he cannot document the exact steps he followed to determine the baseline, because then he would also have to document the steps he took to falsify the baseline. His spreadsheet doesn't even have a column that would show the baseline deaths, because I think if his followers showed that the slope of the baseline took a sharp dive downwards after 2019, they would be asking how it is justified, and why it's not documented anywhere.
His spreadsheet doesn't even show how he determined the lines of his legacy trend and emergent trend. He probably just drew the lines by hand, because for example the starting point of the legacy trend is slightly before the first week of 2018, but if he would've calculated the line by doing a real regression of the numeric data, you'd expect the line to start exactly on the first week of 2018. And similarly the ending points of the legacy trend and emergent trend are not even on the same week.
ES now also added new blue and orange trend lines to the plot for infant mortality from the CDC report, but he seems to have drawn the lines by hand, because their starting and ending points don't match the years on the x-axis, and the slope percentages he added next to the lines don't match the actual slope of the lines:
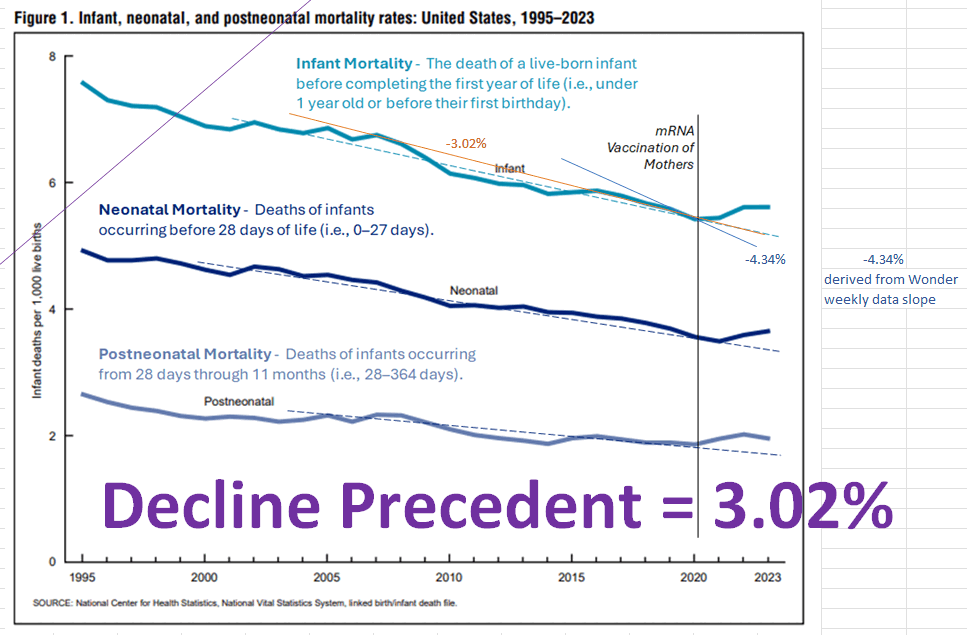
The blue line is supposed to represent an annual decrease of about
4.34%. But the blue line decreased from about 6.29 in 2015 to 5.08 in
2022, which is an annual decrease of only about 2.8% if you divide the
total decrease rate with the number of years:
(1-5.07/6.29)/7 (where by the number of years, I mean the
number of steps from the previous year to the next year, which is the
count of years minus 1). Or the formula for a compound annual growth
rate gives an annual decrease of about 3.0%:
(5.07/6.29)^(1/7)-1.
I guess the 4.34% annual decrease is supposed to represent the slope of his "legacy trend", which decreases from about 431 on week 1 of 2018 to 312 on week 1 of 2025, so it's an approximately 3.9% decrease if you divide the total decrease rate by the number of years, or an approximately 4.5% decrease if you use the formula for CAGR.
So even though the blue line in the spreadsheet is clearly too steep relative to the real pre-COVID trend, the legacy trend is even steeper, which shows how the pre-COVID rate of decline is exaggeated by the legacy trend. And ES falsely portrays the blue line as representing a 4.34% decrease.
The orange line is supposed to decrease by 3.02% per year, but
actually it decreases from about 7.04 in 2004 to 5.27 in 2022, which is
about 1.4% per year if you divide the total decrease rate with the
number of years: (1-5.27/7.04)/18. Or it's about 1.6% with
CAGR: (5.27/7.04)^(1/18)-1.
And even the orange line is steeper than the real 2004-2019 linear trend in the CDC data, where you get a decrease of only about 1.3% per year if you divide the total decrease rate between 2004 and 2022 with the number of years from 2004 to 2022:
# from cdc.gov/nchs/data/nvsr/nvsr74/nvsr74-07.pdf#page=8 d=CJ(year=1995:2023) d$rate=c(757,730,721,719,704,689,684,695,684,678,686,668,675,661,639,614,607,598,596,582,590,587,579,567,558,542,544,561,561)/100 d$trend=predict(lm(rate~year,d[year%in%2004:2019]),d) d[,(1-trend[year==2022]/trend[year==2004])/18] # 0.01282821 (about 1.3%)
The next table shows that the real infant mortality rate in the CDC
report was about 5.42 in 2020, but for some reason ES came up with his
own "normalized" infant mortality rate of
4.54 for 2020. He used his normalized rate to derive his unrealistically
steep figure of a 3.02% annual decline in infant mortality between 2007
and 2020. I think ES calculated the figure of 3.02% by either using the
formula for CAGR ((4.54/6.75)^(1/13)-1, about 3.00%), or as
the difference between the start and end divided by the mean of the
start and end and then divided by the number of years
((6.75-4.54)/((6.75+4.54)/2)/13, about 3.01%). Both of
those were slightly off from 3.02% when I used rounded numbers, but ES
may have used unrounded numbers:
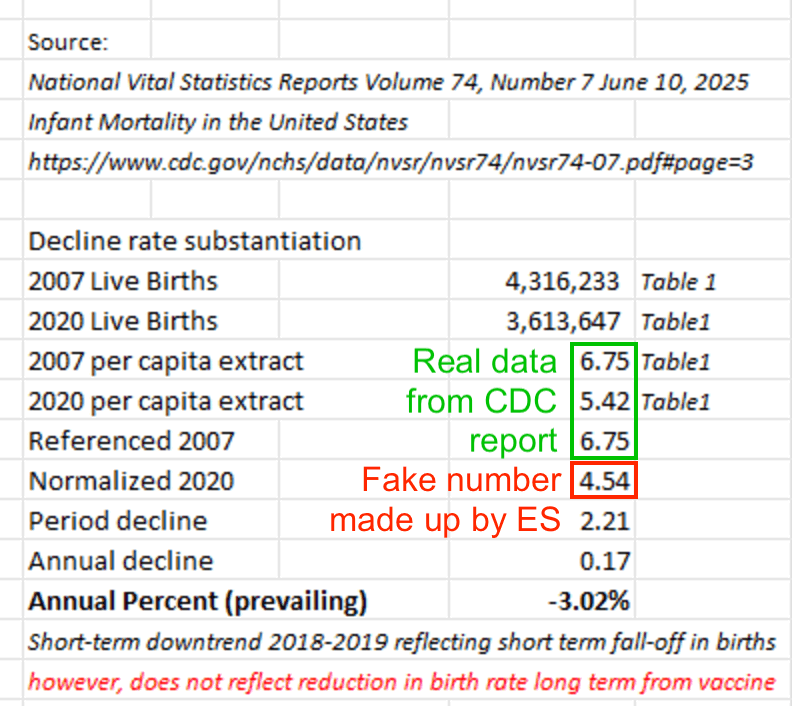
My best guess for how ES derived his "normalized" infant mortality rate is that first he calculated the annual decline percent of his "legacy trend", which was about 4.3%. Then he manually drew a line in the CDC plot that approximated the slope between 2019 and 2020, and he falsely declared that the line decreased by about 4.3% per year. Then he thought that the longer-term rate of decline before COVID looked about a third less steep than his blue line, so he picked a fake infant mortality rate for 2020 that gave him an annual rate of decline of about 3%.
But regardless of how ES determined the normalized rate for 2020, it's ridiculously low compared to the real infant mortality rate in 2020:
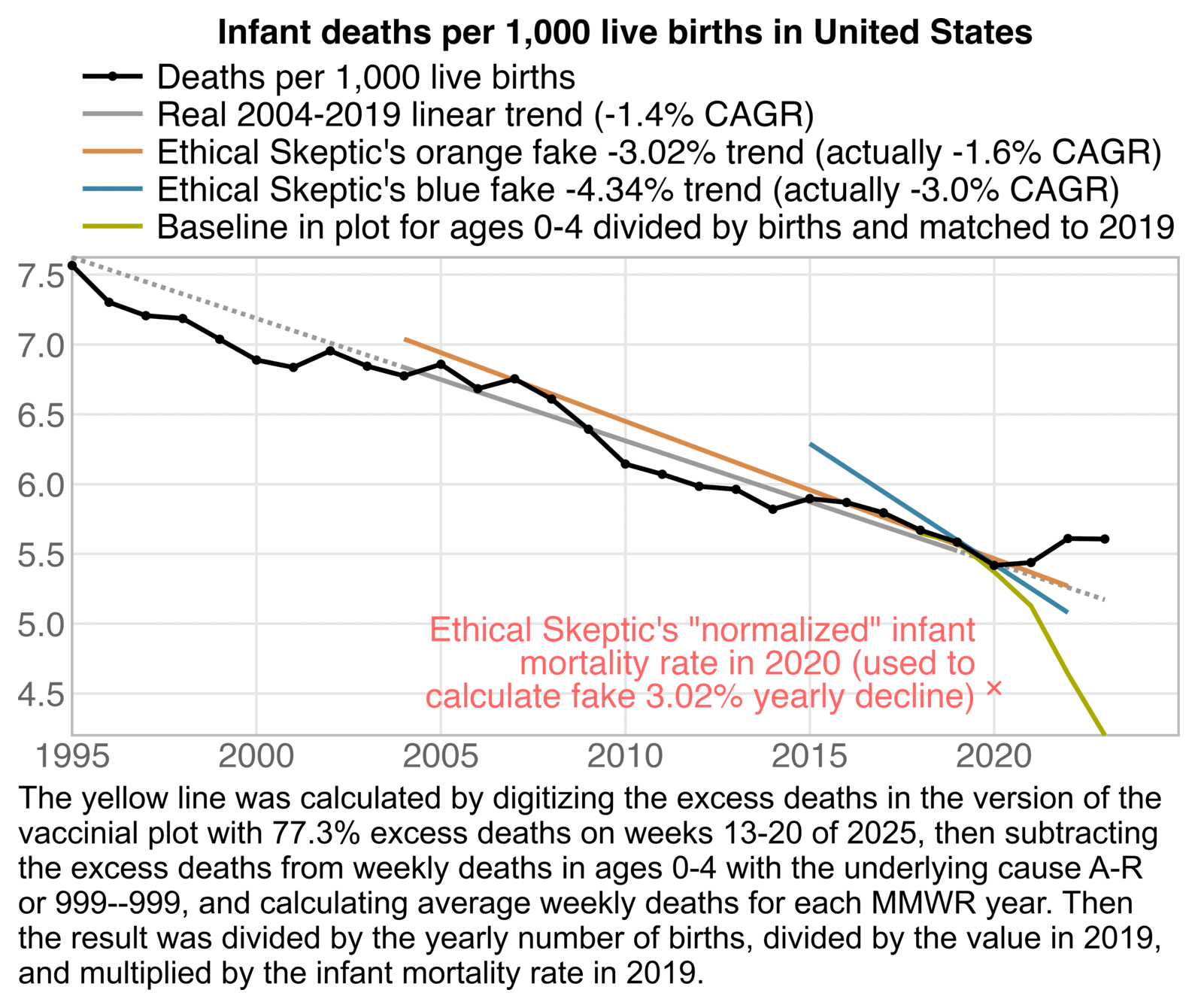
# from cdc.gov/nchs/data/nvsr/nvsr74/nvsr74-07.pdf#page=8
d=CJ(year=1995:2023)
d$born=c(3899589,3891494,3880894,3941553,3959417,4058882,4026036,4021825,4090007,4112055,4138573,4265593,4316233,4247726,4130665,3999386,3953590,3952841,3932181,3988076,3978497,3945875,3855500,3791712,3747540,3613647,3664292,3667758,3596017)
d$dead=c(29505,28419,27968,28325,27865,27961,27523,27970,27995,27860,28384,28509,29153,28075,26408,24572,24001,23654,23446,23211,23458,23157,22341,21498,20927,19578,19928,20577,20162)
d[,rate:=dead/born*1e3]
p1=d[,.(x=year,y=dead/born*1e3,fit=T)]
p1$z="Deaths per 1,000 live births"
p2=copy(p1)[,fit:=x%in%2004:2019]
p2=p2[,.(x,y=predict(lm(y~x,p2[fit==T]),p2),fit)]
p2$z="Real 2004-2019 linear trend (-1.4% CAGR)"
p3=data.table(x=c(2004,2022),y=c(7.04,5.27),fit=T)
cagr=p3[,(y[2]/y[1])^(1/(x[2]-x[1]))-1]
p3$z=sprintf("Ethical Skeptic's orange fake -3.02%% trend (actually %.1f%% CAGR)",cagr*100)
p4=data.table(x=c(2015,2022),y=c(6.29,5.08),fit=T)
cagr=p4[,(y[2]/y[1])^(1/(x[2]-x[1]))-1]
p4$z=sprintf("Ethical Skeptic's blue fake -4.34%% trend (actually %.1f%% CAGR)",cagr*100)
eth=fread("https://sars2.net/f/wondervaccinial0to4.csv")
p5=merge(d,na.omit(eth)[,.(y=mean(dead-excess),fit=T),.(year)])[,.(x=year,fit=T,y=y/rate)]
p5[,y:=y*p1[x==2019,y]/p5[x==2019,y]]
p5$z="Baseline in plot for ages 0-4 divided by births and matched to 2019"
p=rbind(p1,p2,p3,p4,p5)[,z:=factor(z,unique(z))]
xmin=1995;xmax=2025;xbreak=seq(xmin,xmax-5,5)
ybreak=pretty(c(p$y),8);ymin=min(p$y);ymax=max(p$y)
ggplot(p)+
geom_hline(yintercept=0,color="gray75",linewidth=.4)+
annotate("rect",xmin=xmin,xmax=xmax,ymin=ymin,ymax=ymax,linewidth=.4,lineend="square",fill=NA,color="gray75")+
annotate("point",x=2020,y=4.54,color="red",shape=4,size=1.3,stroke=.6)+
annotate("text",x=2019.5,y=4.4,color="red",size=3.87,lineheight=.8,label=str_wrap("Ethical Skeptic's \"normalized\" infant mortality rate in 2020 (used to calculate fake 3.02% yearly decline for orange line)",35),hjust=1,vjust=0)+
geom_line(aes(x,y,color=z),linetype="11",linewidth=.7)+
geom_line(data=p[fit==T],aes(x,y,color=z),linewidth=.7)+
geom_line(data=p[z==z[1]],aes(x,y,color=z),linewidth=.7)+
geom_point(aes(x,y,color=z,alpha=z),size=1.4,stroke=0)+
labs(x=NULL,y=NULL,title="Infant deaths per 1,000 live births in United States")+
scale_x_continuous(limits=c(xmin,xmax),breaks=xbreak)+
scale_y_continuous(limits=c(ymin,ymax),breaks=ybreak,labels=\(x)sprintf("%.1f",x))+
scale_color_manual(values=c("black","gray60","#DA8A48","#3A84A7","#aaaa00"))+
scale_alpha_manual(values=c(1,0,0,0,0))+
coord_cartesian(clip="off",expand=F)+
theme(axis.text=element_text(size=11,color="gray40"),
axis.ticks.length=unit(0,"pt"),
legend.background=element_blank(),
legend.box.spacing=unit(0,"pt"),
legend.direction="vertical",
legend.key=element_blank(),
legend.key.height=unit(12,"pt"),
legend.key.spacing.x=unit(0,"pt"),
legend.key.width=unit(24,"pt"),
legend.margin=margin(-2,,4),
legend.position="top",
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=11,margin=margin(,,,2)),
legend.title=element_blank(),
panel.background=element_blank(),
panel.grid.major=element_line(linewidth=.4,color="gray90"),
plot.title=element_text(size=11,hjust=.5,face=2,margin=margin(,,4)))
ggsave("1.png",width=5.3,height=3.5,dpi=300*4)
This table also shows how ES exaggerated the annual decline percentages of his orange and blue lines:
| Line | X1 | X2 | Y1 | Y2 |
Claimed by ES |
Compound annual growth rate: (Y2/Y1)^(1/(X2-X1))-1 |
Total decrease rate divided by number of years: (Y2/Y1-1)/(X2-X1) |
Difference between start and end divided by mean of start and end divided by number of years: (Y2-Y1)/mean(Y1,Y2)/(X2-X1) |
|---|---|---|---|---|---|---|---|---|
| Real infant mortality rate in the CDC report | 2007 | 2020 | 6.75 | 5.42 | NA | -1.67% | -1.52% | -1.68% |
| 2007 CDC rate with fake normalized rate for 2020 | 2007 | 2020 | 6.75 | 4.54 | -3.02% | -3.00% | -2.52% | -3.01% |
| Orange line | 2004 | 2022 | 7.04 | 5.27 | -3.02% | -1.60% | -1.40% | -1.60% |
| Blue line | 2015 | 2022 | 6.29 | 5.08 | -4.34% | -3.01% | -2.75% | -3.04% |
Ethical Skeptic posted the plots below and wrote: "You completely ignored the 2017 downturn in birth rate (left panel) - which is CONFIRMED by the weekly Wonder data (right panel)." [https://x.com/EthicalSkeptic/status/1970833812050325505]

So as a justification for why his legacy trend should have an extremely steep slope, he said that there had been a decreasing trend in the rate of births before COVID.
His orange line shows excess births relative to a 2014-2023 trend in California. It's confusing how his plot doesn't show the baseline and actual births but only the excess births. In the years 2014-2016 there was in fact a fairly high number of births relative to the years before 2014 or the years after 2016, but the drop in births after 2016 wasn't steep enough to justify the extremely steep "legacy trend".
His plot for California indicates that in 2022 when the excess number of births was only slightly below 0%, it was because of a "temporary lockdown push-off effect". However in the next plot when I used a 2014-2023 baseline like ES, but I looked at births in the whole United States and not only California, and I didn't apply his adjustments, I got positive excess births each year in 2022, 2023, and 2024. So there rather seems to have been a temporary reduction in births in 2020 and 2021, but by 2022-2024 the births had returned back to the pre-COVID trend (or the births were either slightly above or slightly below the trend depending on how you calculate the trend). And the age-standardized fertility rate was above the 2014-2023 trend even in 2021:
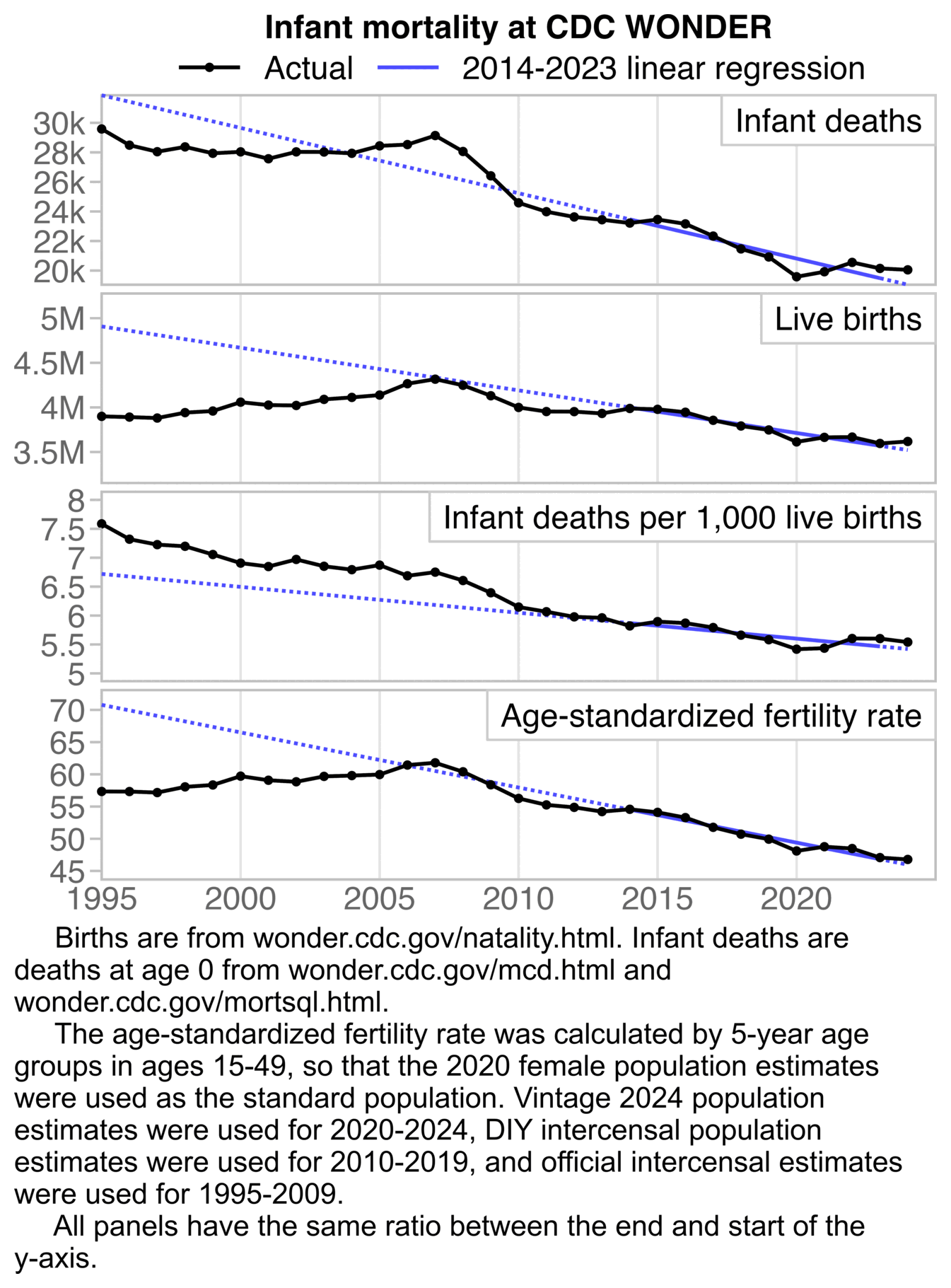
t=fread("https://sars2.net/f/wonderbornmaternalage.csv")
dlf=\(x,y=sub(".*/","",x),...)for(i in 1:length(x))download.file(x[i],y[i],quiet=T,...)
files=c("1990-2000/intercensal/national/us-est90int-07.csv",
"2000-2010/intercensal/national/us-est00int-alldata.csv",
"2010-2020/national/asrh/nc-est2020-agesex-res.csv",
"2020-2024/national/asrh/nc-est2024-agesex-res.csv")
dlf(paste0("https://www2.census.gov/programs-surveys/popest/datasets/",files))
t1=fread("us-est90int-07.csv",fill=T,skip=2)[V1%like%"July"&V2!="All Age"]
t1[,age:=as.integer(sub("\\+","",V2))]
d1=t1[,.(pop=sum(V5)),.(age,year=as.integer(sub(".* ","",V1)))]
t2=fread("us-est00int-alldata.csv")
d2=t2[YEAR!=2010&AGE<85&MONTH==7,.(pop=sum(TOT_FEMALE)),.(year=YEAR,age=AGE)]
t3=fread("nc-est2020-agesex-res.csv")
d3=t3[SEX==2&AGE!=999,.(age=AGE,pop=unlist(.SD[,-(1:4)]),year=rep(2010:2020,each=.N))]
t4=fread("nc-est2024-agesex-res.csv")
d4=t4[SEX==2&AGE!=999,.(age=AGE,pop=unlist(.SD[,-(1:3)]),year=rep(2020:2024,each=.N))]
mult=d4[year==2020,pop]/d3[year==2020,pop]
d3=d3[year<2020,{x=(year-2010)/10;.(age,pop=(mult[age+1]*x+1-x)*pop,year)}]
a=t[,.(born=sum(born)),.(year,age)]
a=merge(a,rbind(d1,d2,d3,d4)[age%in%15:49,.(pop=sum(pop)),.(age=age%/%5*5,year)])
asfr=a[,.(y=sum(born/pop*a[year==2020,pop/sum(pop)][factor(age)]*1e3)),.(x=year)]
t=fread("https://sars2.net/f/wondernatality.csv")
t=t[,.(born=sum(born)),year]
t=rbind(t,list(1995:2002,c(3899589,3891494,3880894,3941553,3959417,4058814,4025933,4021726)))
d=fread("https://sars2.net/f/uspopdead.csv")[age==0,.(year,dead)]
d=rbind(d,list(1995:1998,c(29583,28487,28045,28371)))
p=merge(t,d)[year<2025,.(x=year,y=c(dead,born,dead/born*1e3),facet=rep(1:3,each=.N))]
p=rbind(p,asfr[,facet:=4])
p1=p[,fit:=1][,z:=1]
p2=copy(p1)[,fit:=x%in%2014:2023]
p2=p2[,.(x,fit,y=predict(lm(y~x,.SD[fit==T]),.SD),z=2),facet]
p=rbind(p1,p2)
p[,facet:=factor(facet,,c("Infant deaths","Live births","Infant deaths per 1,000 live births","Age-standardized fertility rate"))]
p[,z:=factor(z,,c("Actual","2014-2023 linear regression"))]
xstart=1995;xend=2025;xbreak=seq(xstart,xend-1,5)
p=p[x%in%xstart:xend]
ylim=p[,.(min=min(y),max=max(y)),facet]
rat=ylim[,max(max/min)]
ylim=ylim[,{x=(rat*min-max)/(1+rat);.(min=min-x,max=max+x,facet)}]
ggplot(p)+
facet_wrap(~facet,ncol=1,dir="v",scales="free_y")+
geom_vline(xintercept=xbreak,color="gray90",linewidth=.4)+
geom_rect(data=ylim,aes(ymin=min,ymax=max),xmin=xstart,xmax=xend,lineend="square",linejoin="mitre",fill=NA,color="gray75",linewidth=.4)+
geom_line(aes(x,y,color=z),linetype="11",linewidth=.6)+
geom_line(data=p[fit==T],aes(x,y,color=z),linewidth=.6)+
geom_line(data=p[z==z[1]],aes(x,y,color=z),linewidth=.6)+
geom_point(aes(x,y,alpha=z),stroke=0,size=1.5)+
geom_label(data=ylim,aes(xend,max,label=facet),vjust=1,hjust=1,label.r=unit(0,"pt"),label.padding=unit(4.5,"pt"),label.size=.4,size=3.87,fill="white",color="gray80")+
geom_label(data=ylim,aes(xend,max,label=facet),vjust=1,hjust=1,label.r=unit(0,"pt"),label.padding=unit(4.5,"pt"),label.size=0,size=3.87,fill=NA)+
labs(title="Infant mortality at CDC WONDER",x=NULL,y=NULL)+
scale_x_continuous(limits=c(xstart,xend),breaks=xbreak)+
scale_y_continuous(breaks=pretty,labels=\(x)ifelse(x>=1e3,ifelse(x>=1e6,paste0(x/1e6,"M"),paste0(x/1e3,"k")),x))+
scale_color_manual(values=c("black",hsv(2/3,.7,1)))+
scale_alpha_manual(values=c(1,0,0))+
coord_cartesian(clip="off",expand=F)+
theme(axis.text=element_text(size=11,color="gray40"),
axis.text.y=element_text(margin=margin(,1.5)),
axis.ticks=element_line(linewidth=.4,color="gray75"),
axis.ticks.length.x=unit(0,"pt"),
axis.ticks.length.y=unit(4,"pt"),
legend.background=element_blank(),
legend.box.spacing=unit(0,"pt"),
legend.direction="horizontal",
legend.key=element_blank(),
legend.key.height=unit(11,"pt"),
legend.key.width=unit(26,"pt"),
legend.margin=margin(,,4),
legend.position="top",
legend.spacing.x=unit(2,"pt"),
legend.spacing.y=unit(0,"pt"),
legend.text=element_text(size=11,vjust=.5),
legend.title=element_blank(),
panel.background=element_blank(),
panel.grid=element_blank(),
panel.spacing=unit(3,"pt"),
plot.title=element_text(size=11,hjust=.5,face="bold",margin=margin(,,2)),
strip.background=element_blank(),
strip.text=element_blank())
ggsave("1.png",width=4.5,height=4.4,dpi=300*4)
But anyway, regardless of whether you look at infant deaths, live births, infant mortality rate, or age-standardized fertility rate, in the plot above the pre-COVID trend is not as steep as a 2018-2020 trend, because there was an unusually low number of both births and infant deaths in 2020. So 2020 should not be included in the fitting period of the baseline.