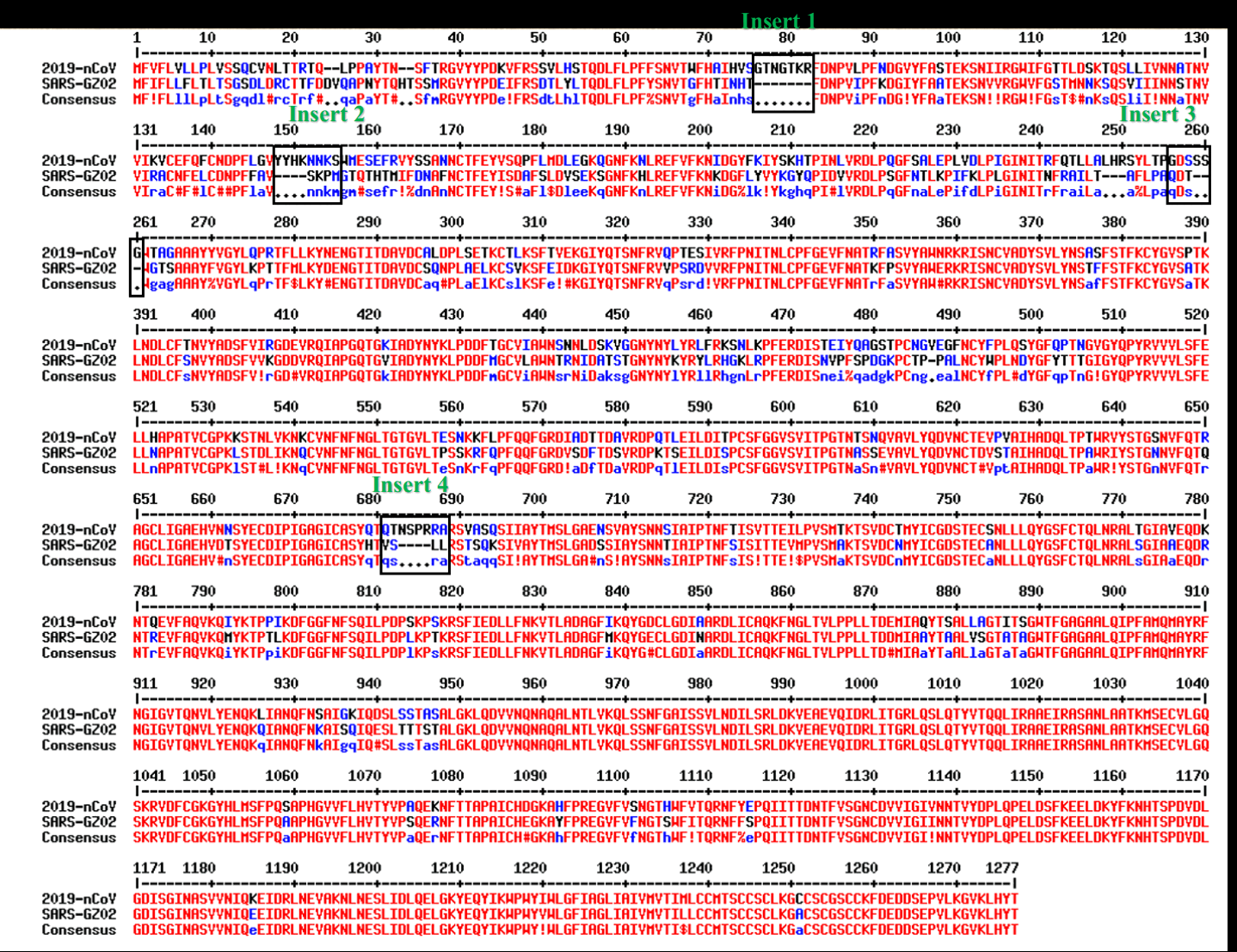
Pradhan et al. did a pairwise alignment of the spike protein of SARS2 against SARS1, and they identified 4 regions where SARS2 had an insert relative to SARS1: [https://www.researchgate.net/publication/338957445_Uncanny_similarity_of_unique_inserts_in_the_2019-nCoV_spike_protein_to_HIV-1_gp120_and_Gag]

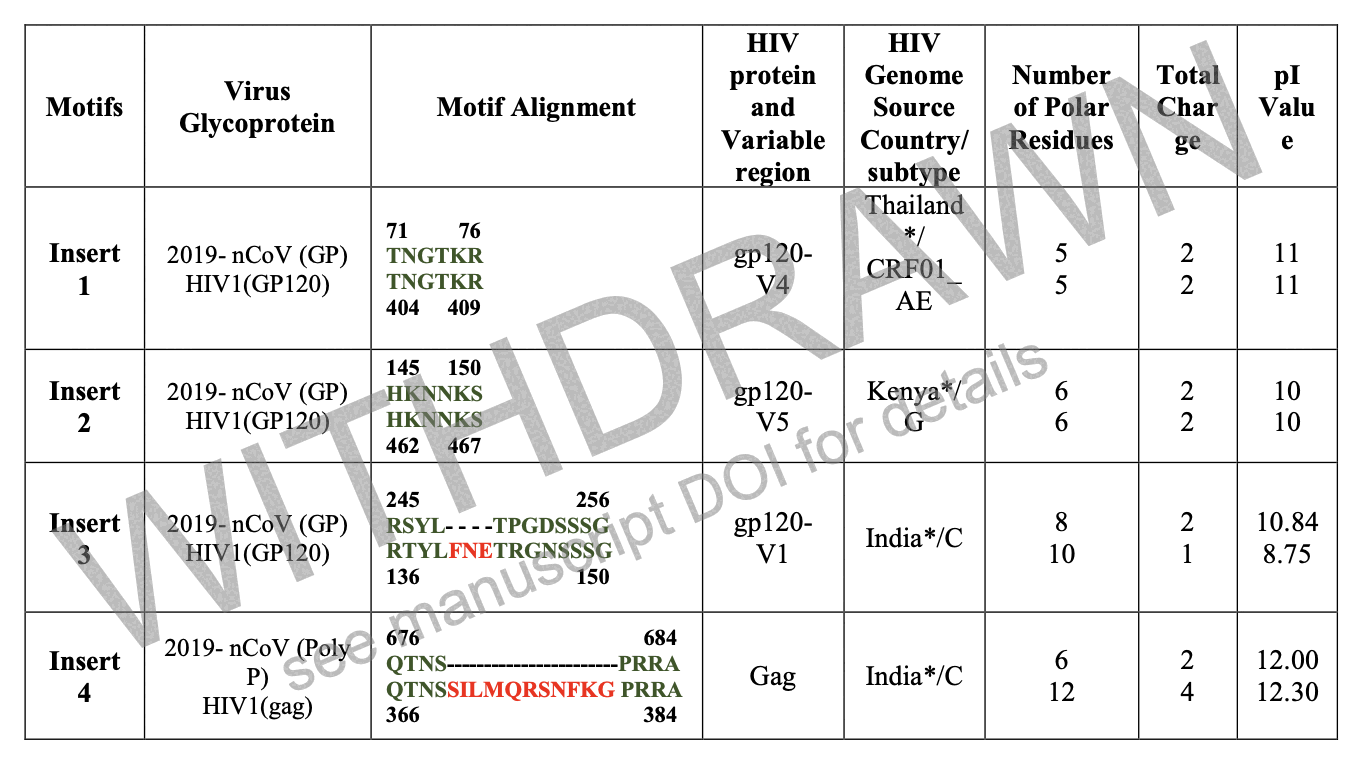
When Pradhan et al. did BLAST searches for the inserts and the surrounding amino acids, they found that for example the last 6 aa of the first 7-aa insert had a perfect match to an HIV sequence from Thailand, and the last 2 aa of the second insert along with the next 4 aa had a perfect match to an HIV sequence from Kenya:
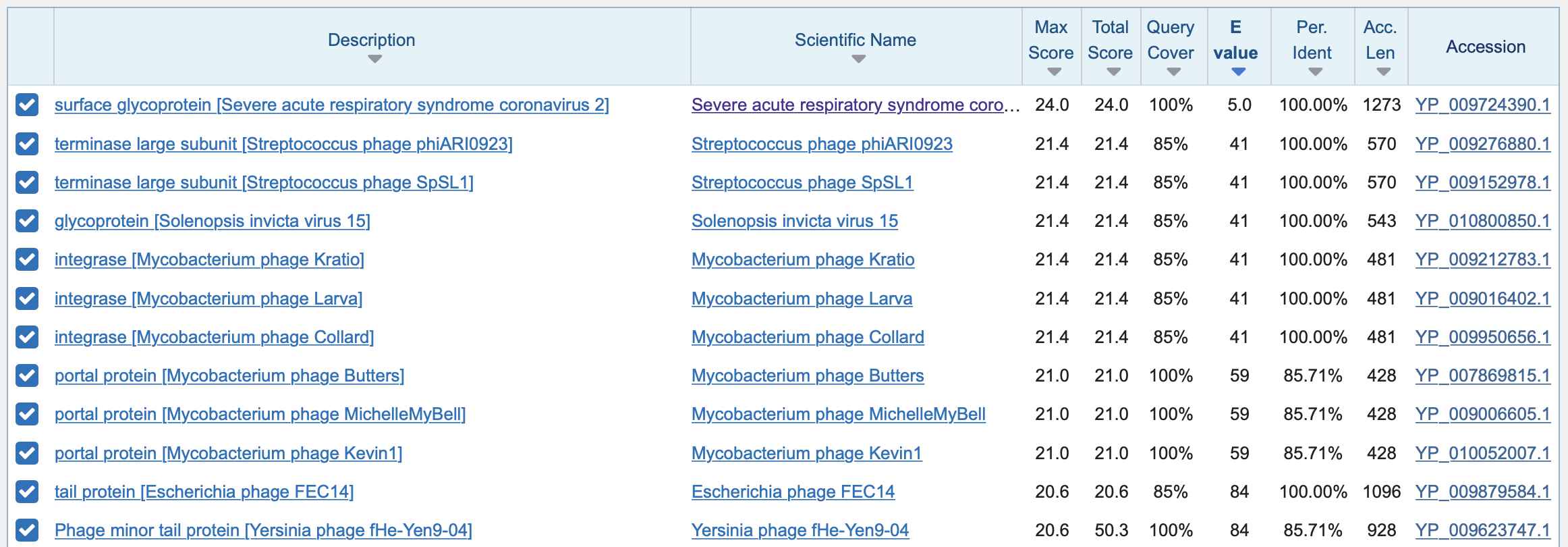
I tried searching for the first 7-aa insert in protein BLAST:
https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch.
I entered GTNGTKR to the text field at the top, I set the
database to refseq_protein, I set the organism as "Viruses (taxid:10239)", and I clicked BLAST.
There were many different bacteriopages and a few non-phage viruses which matched 6 out of 7 aa so that one aa at either end was clipped by the local alignment, so that there was 100% identity but 85% query coverage:
HIV-1 was not included in the results above, because I searched in a database which only contained refseq proteins. The standard reference genome of HIV-1 is called HXB2, and its envelope protein matches only 3 out of 7 aa of Pradhan's first insert:
$ curl 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta_cds_aa&id=NC_001802'>hiv.aa $ seqkit locate -p GTNGTKR -m3 <(seqkit grep -nrp gene=env hiv.aa)|cut -f3,5-|column -t # no matches for max mismatch 3 pattern start end matched $ seqkit locate -p GTNGTKR -m4 <(seqkit grep -nrp gene=env hiv.aa)|cut -f3,5-|column -t # 7 matches for max mismatch 4 pattern start end matched GTNGTKR 232 238 TFNGTGP GTNGTKR 300 306 NNNTRKR GTNGTKR 321 327 GKIGNMR GTNGTKR 354 360 GNNKTII GTNGTKR 404 410 GSNNTEG GTNGTKR 495 501 GVAPTKA GTNGTKR 822 828 VAEGTDR
Next I tried doing another BLAST search for Pradhan's first insert, but I set the "Non-redundant protein sequences (nr)" database and I set the organism to HIV-1. There were 3 different spots of the envelope protein which all matched 6/7 aa of the insert so that one aa at either end was cut off:
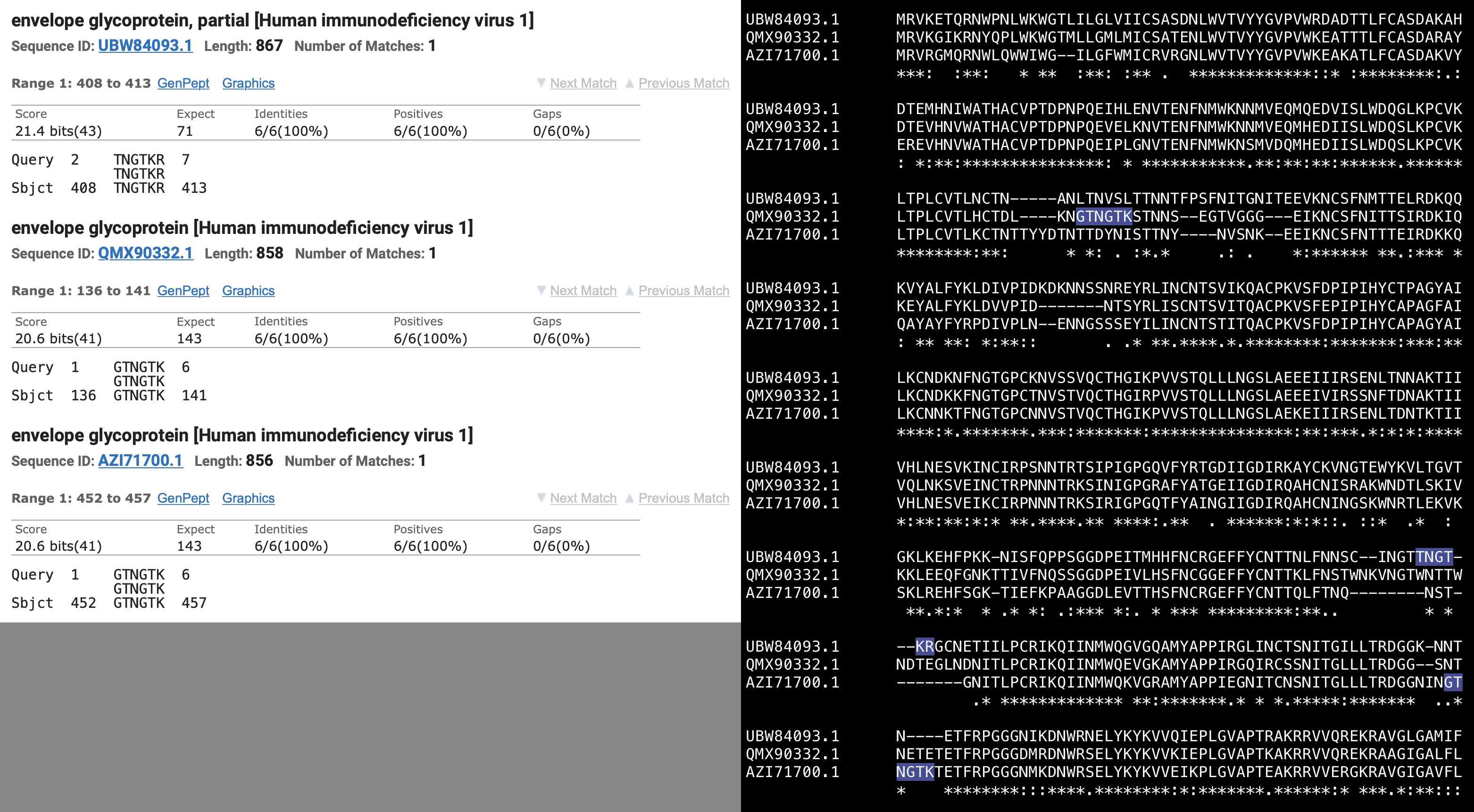
There's more than a million sequences of HIV-1 in GenBank: https://www.ncbi.nlm.nih.gov/nuccore/?term=%22Human+immunodeficiency+virus+1%22%5Bporgn%3A%5f%5ftxid11676%5D. I don't know how many of them are included at BLAST, but in any case it's not that unusual that a handful of sequences would randomly happen to match 6 out of 7 aa of Pradhan's first insert.
The env protein of HIV is divided into the gp120 and gp41
polyproteins, but Pradhan et al.'s first insert matched the gp120 part.
I downloaded 125,046 gp120 sequences from the HIV sequence database of
the Los Alamos National Laboratory:
https://www.hiv.lanl.gov/components/sequence/HIV/search/search.html.
None of them matched the full GTNGTKR insert, but there
were 15 sequences that matched the first 6 aa and there were 12
sequences which matched the last 6 aa, and the matches were at 3
different spots:
$ seqkit stat HIV-1_env.fasta|column -t file format type num_seqs sum_len min_len avg_len max_len HIV-1_env.fasta FASTA Protein 125,046 105,184,828 479 841.2 1,469 $ seqkit locate -p GTNGTK HIV-1_env.fasta|cut -f1,5-7|column -t seqID start end matched C.IN.-.VB49.EF694035 135 140 GTNGTK C.ZA.2009.CAP177_39mo_30.KC833437 452 457 GTNGTK C.ZA.2009.CAP177_4260_186wpi_plasma_12.MK205618 452 457 GTNGTK C.ZA.2009.CAP177_4260_186wpi_cvl_73.MK205633 452 457 GTNGTK B.US.1990.1580m.11a.MT861903 136 141 GTNGTK B.US.1990.1580m.13s.MT861906 136 141 GTNGTK B.US.1990.1580m.15.MT861908 136 141 GTNGTK B.US.1990.1580m.16a.MT861910 136 141 GTNGTK B.US.1990.1580m.17.MT861911 136 141 GTNGTK B.US.1990.1580m.18.MT861913 136 141 GTNGTK B.US.1990.1580m.22.MT861919 136 141 GTNGTK B.US.1990.1580m.3a.MT861922 136 141 GTNGTK B.US.1990.1580m.5.MT861925 136 141 GTNGTK B.US.1990.1580m.7.MT861927 136 141 GTNGTK A1CD.TZ.2012.30196v23_env12.OM825465 460 465 GTNGTK $ seqkit locate -p TNGTKR HIV-1_env.fasta|cut -f1,5-7|column -t seqID start end matched 01_AE.TH.2006.AA042a12R.JX447199 364 369 TNGTKR 01_AE.TH.2006.AA042a13R.JX447200 399 404 TNGTKR 01_AE.TH.2006.AA042a09R.JX447201 398 403 TNGTKR 01_AE.TH.2006.AA042a08R.JX447202 398 403 TNGTKR 01_AE.TH.2006.AA042a10R.JX447203 404 409 TNGTKR 01_AE.TH.2006.AA042a11R.JX447204 404 409 TNGTKR 01_AE.TH.2006.AA042a02R.JX447205 398 403 TNGTKR 01_AE.TH.2006.AA042a05R.JX447206 398 403 TNGTKR 01_AE.TH.2006.AA042a07R.JX447207 398 403 TNGTKR 01_AE.TH.2006.AA042a03R.JX447208 399 404 TNGTKR 01_AE.TH.2006.AA042a06R.JX447209 404 409 TNGTKR 01_AE.TH.2007.AA042d_ENV24.MZ347079 408 413 TNGTKR
So again it's not that unusual that 29 out of over a 100,000 sequences would happen to match 6 out of 7 aa. The reason why all four of Pradhan's inserts happened to match HIV and not some other virus might simply be that BLAST has a very large number of HIV sequences relative to other viruses.
Pradhan's second so-called insert matches the last 2 aa of the
YYHK insert here and the next 4 aa:

But there's a total of 7 possible 6 aa segments which include at least 2 aa of the 4 aa insert, so again it's not that surprising that one of them would happen to match one out of hundreds of thousands or millions of HIV sequences at BLAST. The gp120 sequences I downloaded had a perfect match for only one out of the 7 segments:
$ x=FLGVYYHKNNKS;for i in {0..6};do seqkit locate -ip ${x:i:6} HIV-1_env.fasta;done
seqID patternName pattern strand start end matched
seqID patternName pattern strand start end matched
seqID patternName pattern strand start end matched
seqID patternName pattern strand start end matched
seqID patternName pattern strand start end matched
seqID patternName pattern strand start end matched
seqID patternName pattern strand start end matched
G.KE.2006.06KE275457V6.KT022379 HKNNKS hknnks + 462 467 hknnks
I also tried downloading all protein sequences of influenza A from the NCBI's influenza virus database. I simply kept the default settings here and clicked "add query": https://www.ncbi.nlm.nih.gov/genomes/FLU/Database/nph-select.cgi#mainform. There were a bit over a million sequences:
$ seqkit stat influenzamega.fa file format type num_seqs sum_len min_len avg_len max_len influenzamega.fa FASTA Protein 1,234,340 497,828,616 1 403.3 775
The spike protein of Wuhan-Hu-1 is 1273 aa long so it has 1268 possible 6 aa subsegments. However 269 of them had an exact match to one or more of the protein sequences of influenza A:
$ curl 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta_cds_aa&id=MN908947'|seqkit grep -nrp gene=S>sars2spike.aa
$ seqkit seq -s sars2spike.aa|awk '{for(i=1;i<length-5;i++)print substr($0,i,6)}'>kmer
$ grep -f kmer <(seqkit seq -s influenzamega.fa)>temp
$ for x in `<kmer`;do grep -m1 $l temp;done|wc -l
269
And in a FASTA file with 125,046 gp120 sequences of HIV, 53 out of 1268 6-mers had a perfect match (but the number would've been bigger if I was looking at all proteins of HIV and not just gp120):
$ grep -fkmer <(seqkit seq -s HIV-1_env.fasta)>temp2 $ for x in `<kmer`;do grep -m1 $x temp2;done|wc -l 53
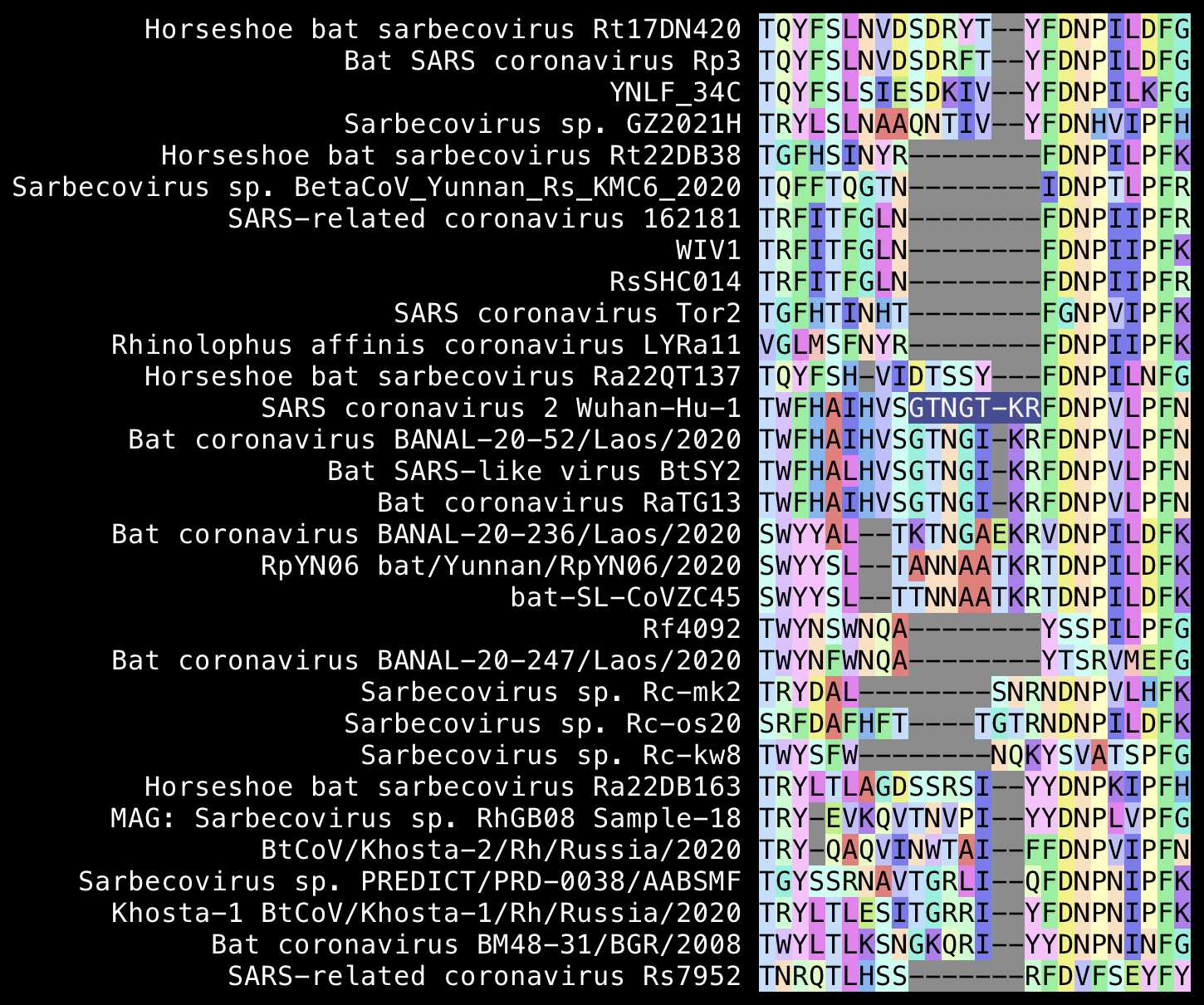
Pradhan's first insert was GTNGTKR, but Wuhan-Hu-1 only
has one or 2 inserted aa in that region relative to European and African
sarbecoviruses, so it rather looks like SARS1 has deletions in that
region. The TKR at the end of the first so-called insert is
also matched by ZC45 and RpYN06:
One of the major flaws in Pradhan's paper was that he didn't report the E-values of the matches, which would've shown how likely similarly close matches were to occur by chance.
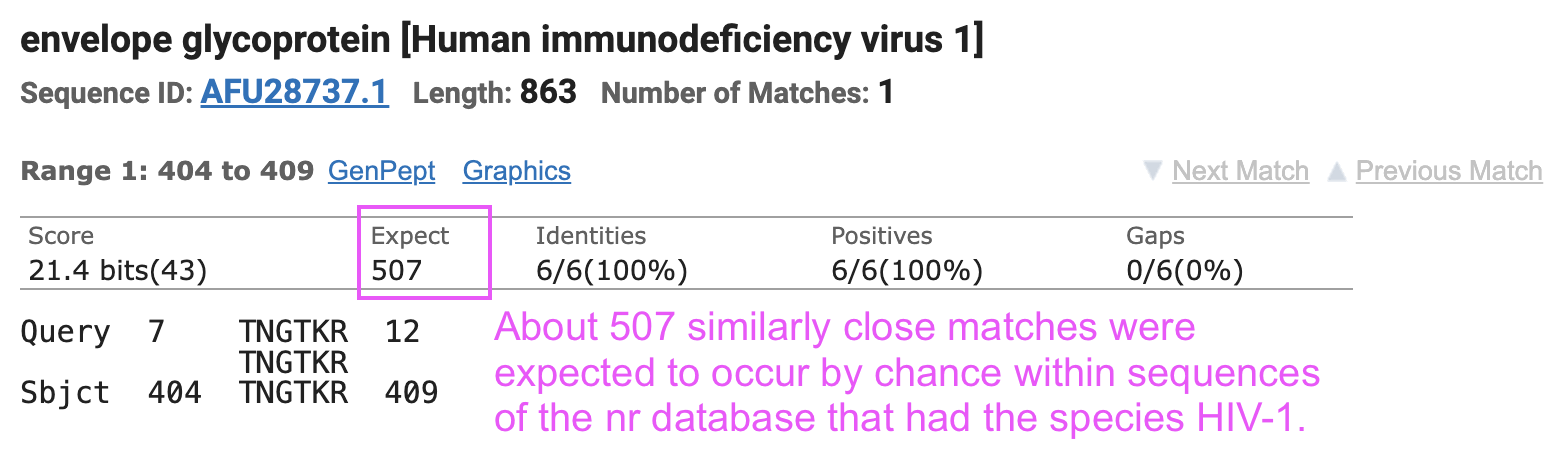
I tried doing a BLAST search for Pradhan's first so-called insert together with the 5 surrounding amino acids on either side, so that kept the default nr database but I restricted the species to HIV-1: https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins.
The matches to the Thai sequences that were listed in Pradhan's paper had an E-value of about 507, which means that about 507 similarly close matches were expected to occur by chance:
There were 97 results which had a better match to the query than Pradhan's Thai sequences. But the lowest E-value among the matches was still 11, so none of the matches were even close to reaching the significance level of 0.05:
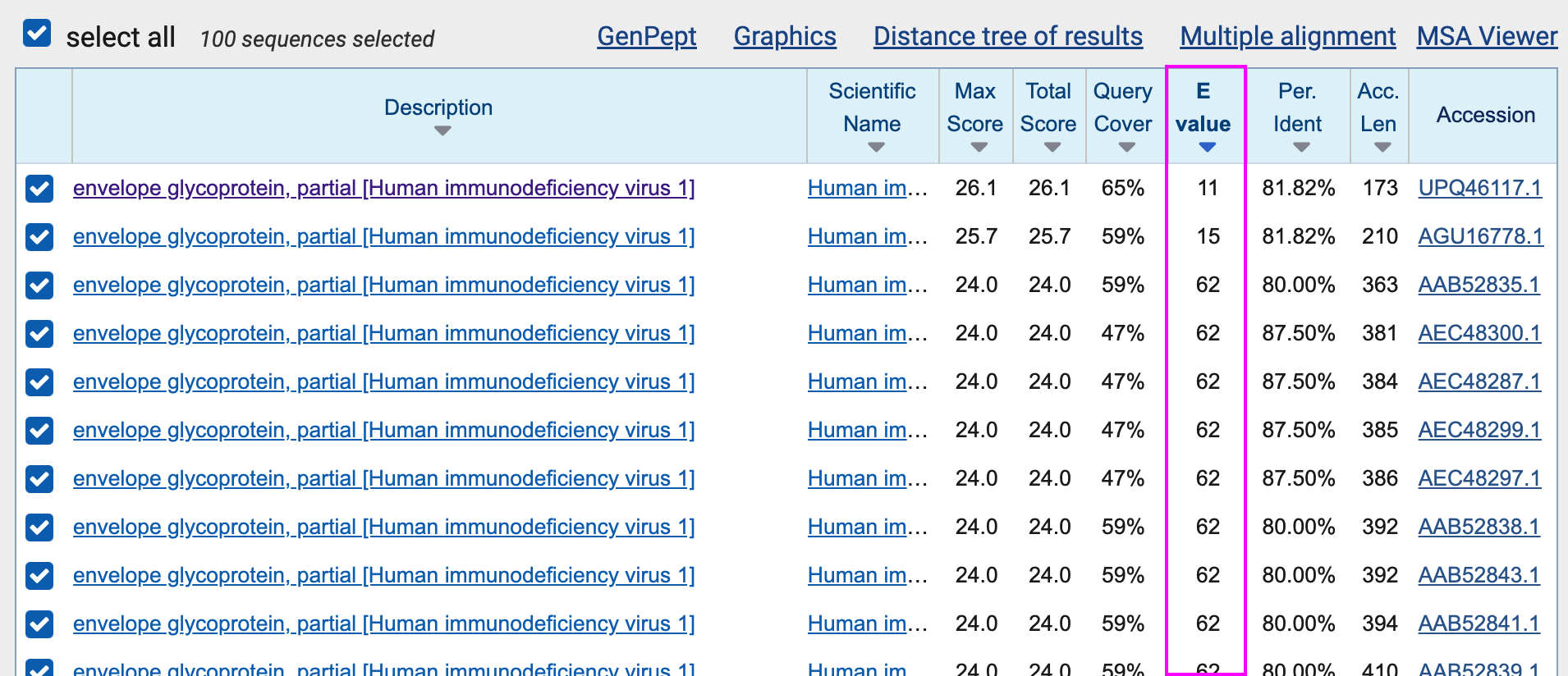
One of the terms which is used to calculate the E-value is the length of the database in letters, so the E-value depends linearly on the length of the database. My database of HIV-1 sequences was about 300 million letters long. If I would've broadened the search to all viruses except SARS-CoV-2, then the length of the database would've been about 7 times bigger, so the E-values would've also been about 7 times higher.
In Pradhan's pairwise alignment SARS2 had the insert
YYHK relative to Tor2. Pradhan's second so-called insert
was HKNNKS, which consisted of the last two aa of
YYHK and the next 4 aa. However the YYHKNNK
without the K in the middle is also included in ZC45 and
ZXC21 (so it's weird that Pradhan didn't mention them since they were
the closest published relatives of SARS2 in January 2020):
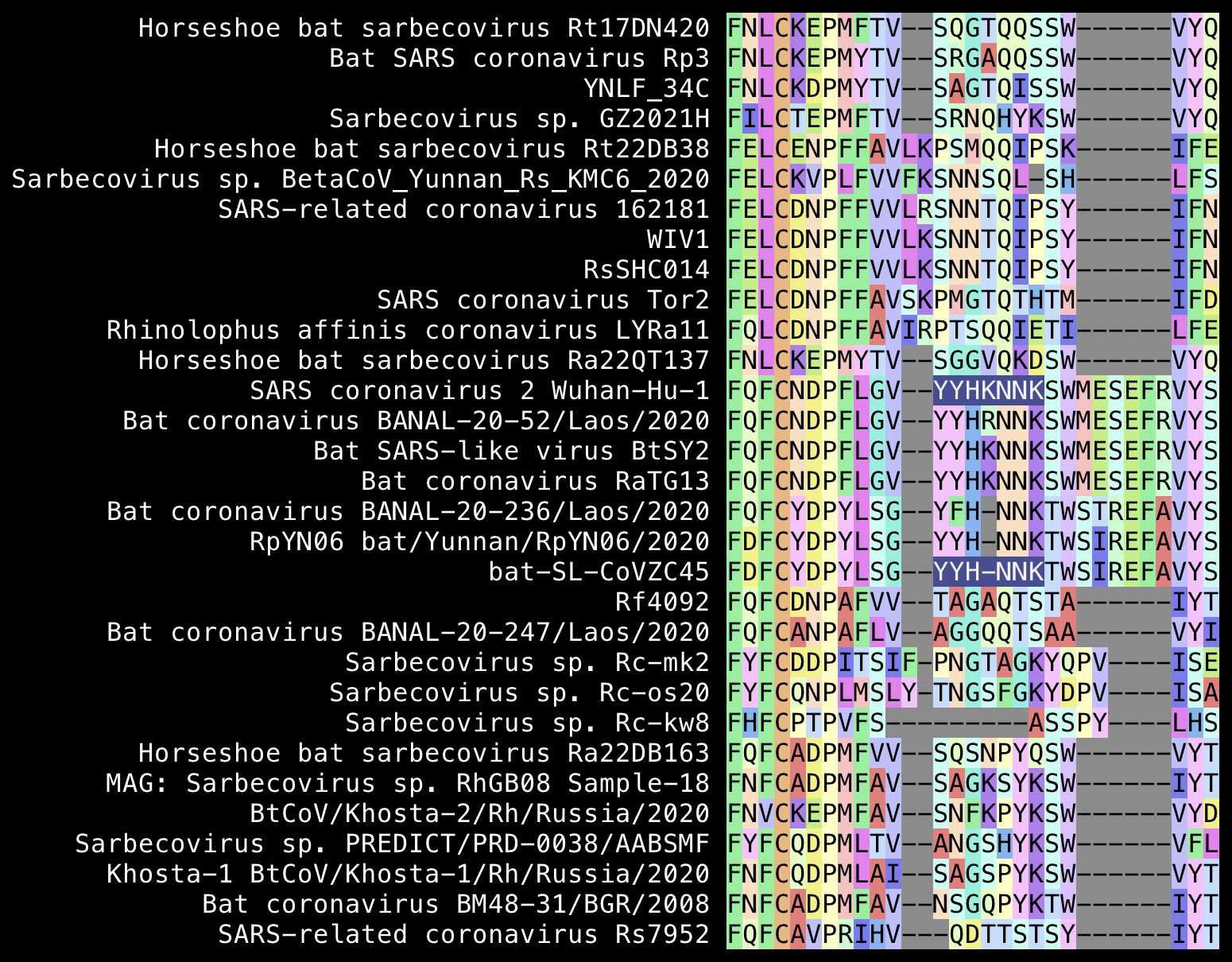
Also from my alignment above it looks that relative to SARS1, SARS2
has the deletion SK and the insertion MESEFR.
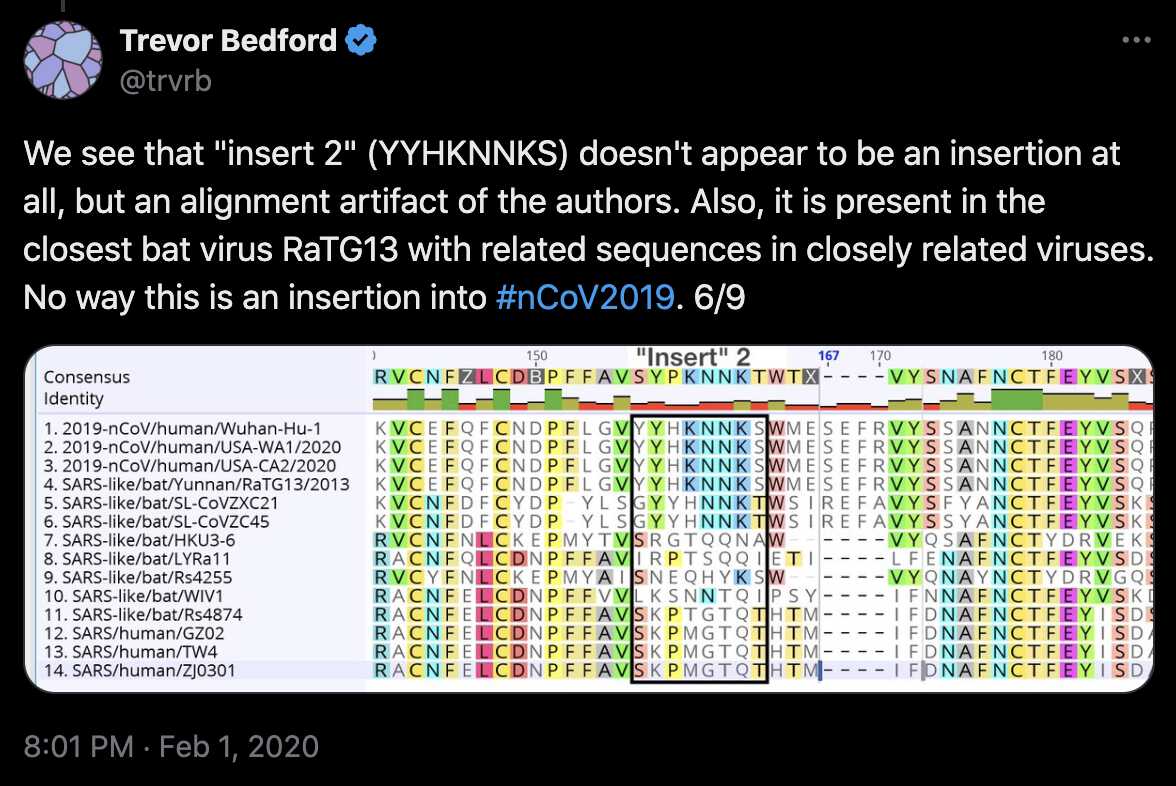
Pradhan et al. identified the locations of their inserts by doing a
pairwise alignment of SARS2 and SARS1, but they would've been able to
identify the locations more accurately if they would've done a multiple
sequence alignment that also included other sarbecoviruses. Trevor
Bedford also pointed out that the YYHK wasn't really even
an insert relative to other sarbecoviruses, but from his alignment it
looks like SARS2 has the insertion SEFR relative to SARS1:
[https://x.com/trvrb/status/1223667730911354880]
The third insert that was highlighted in Pradhan's alignment
consisted of the amino acids SSG, even though a bit before
it there was another 3 aa insert that was not highlighted. But the third
insert might have been placed in the wrong spot because Pradhan et
al. only did a pairwise alignment of SARS2 and SARS1. When I did an
alignment of more viruses, I got a single 6 aa insert in SARS2 relative
to SARS1 instead of two different 3 aa inserts. And my alignment also
shows that the G at the end of SSG is also
included in ZC45 and RpYN06 (and the middle S in
SSG is even included in European and African
sarbecoviruses, even though it might be a coincidence):
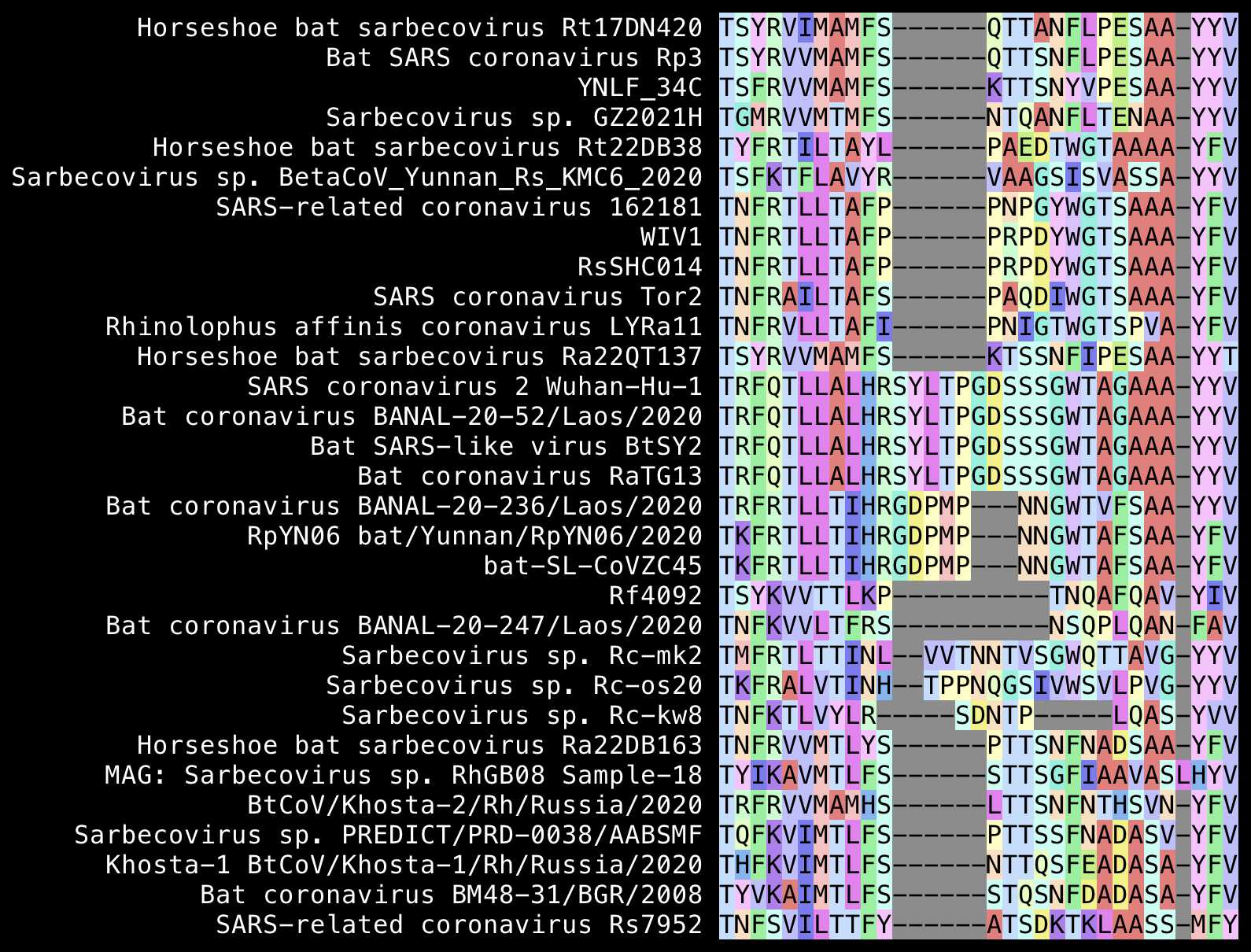
The insert between inserts 2 and 3 that wasn't highlighted by Pradhan
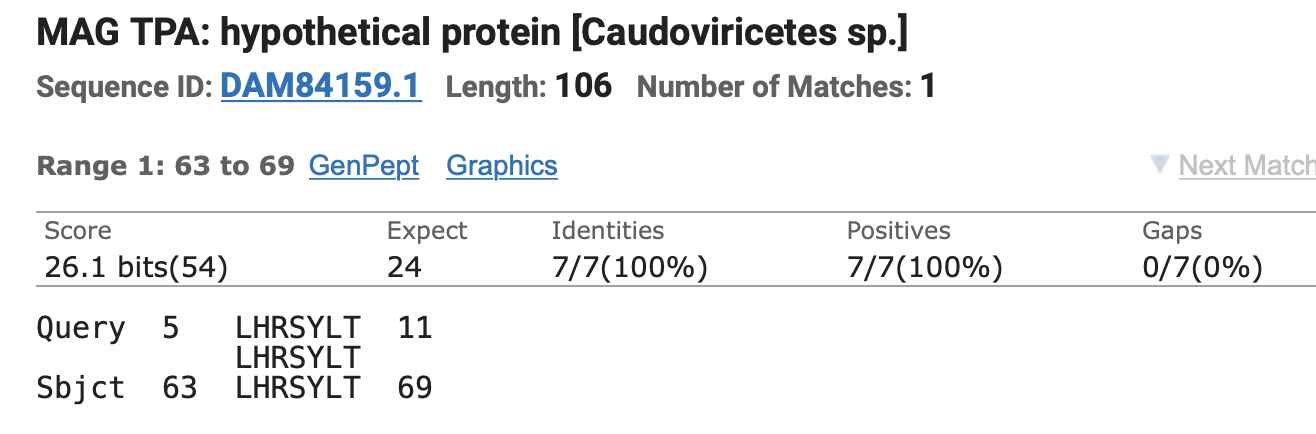
consisted of the amino acids LHR. When I tried searching
for the insert and the surrounding amino acids on BLAST, I found that
the LHR and the next 4 amino acids were matched by a
bacteriophage from the class Caudoviricetes (so it's even
better than Pradhan's first two inserts which only matched 6 amino
acids):
Pradhan's fourth insert consisted of QTNS followed by 11
aa with no match in SARS2 followed by PRRA, which was found
in a single random subtype B sequence of HIV from India. But if the
designers of SARS2 were inspired to borrow the PRRA motif
from HIV, how did they know to look at a random HIV sequence from India
out of hundreds of thousands of published sequences? Here the Indian
sequence is compared to the HXB2 refseq, which contains
QVTNS instead of QTNS, but which only matches
1 out of 4 aa of the PRRA:
$ curl 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=protein&rettype=fasta&id=AKR75206,NP_057850'|mafft --clustalout -
[...]
AKR75206.1 RVLAEAMSQ-TNS-SILMQRSNFKGPRRAVKCFNCGREGHIAKNCRAPRKKGCWKCGKEG
NP_057850.1 RVLAEAMSQVTNSATIMMQRGNFRNQRKIVKCFNCGKEGHTARNCRAPRKKGCWKCGKEG
********* *** :*:***.**:. *: *******:*** *:*****************
^^^^^^^^^^^^^^^^^^^^^
Pradhan insert 4
[...]
Here's code for making amino acid alignments similar to the previous sections of this file:
# download whole genome sequences of sarbecoviruses
curl -sL sars2.net/f/sarbe.fa.xz|xz -dc>sarbe.fa
# efetch multiple sequences with accessions from STDIN
emu()(curl -sd "id=$(paste -sd,)" "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=${1:-nuccore}&rettype=${2:-fasta}&retmode=$3")
# download spike protein sequences of sarbecoviruses and rename them to have same the defline as the whole genome sequences
seqkit seq -ni sarbe.fa|emu '' fasta_cds_aa|seqkit grep -nrp gene=S,spike,surface|sed 's/_prot_.*//;s/lcl|//'|seqkit fx2tab|awk -F\\t 'NR==FNR{a[$1]=$2;next}{print">"$1" "a[$1]"\n"$2}' <(seqkit seq -n sarbe.fa|sed $'s/ /\t/') ->spike.aa
# make a TSV matrix for percentage identity between spike protein sequences
aln()(\mafft --thread 3 --quiet "${@--}")
curl -Ls sars2.net/f/pid.cpp>pid.cpp;g++ pid.cpp -O3 -o pid
seqkit replace -isp '[^X]' -r '' spike.aa|seqkit seq -m 5|seqkit seq -ni|seqkit grep -vf- spike.aa|aln|./pid>spike.pid
# thin out a TSV percentage identity matrix by removing rows with over n% identity to any previous row (default 99%)
thin()(awk -F\\t 'NR>1{for(i=2;i<NR;i++)if($i>x)next;print$1}' x="${1-99}" "${@:2}")
# print color scheme
aacolor()(printf %s\\n A:242:121:121 C:242:182:121 D:242:242:121 G:182:242:121 F:121:242:151 E:121:242:222 T:121:182:242 I:121:121:242 K:182:121:242 L:242:121:242 M:255:191:191 N:255:223:191 P:255:255:191 Q:223:255:191 R:191:255:207 S:191:255:244 H:191:223:255 V:191:191:255 W:223:191:255 Y:255:191:255 -:140:140:140 X:255:255:255 \*:255:255:255|tr : \ )
# display alignment with colorized amino acids
seqcol()(seqkit seq -u|seqkit fx2tab|gawk -F\\t 'NR==FNR{a[$1]=$2;next}{name[FNR]=$1;split($2,z,"");for(i in z)seq[FNR][i]=z[i];if(length($1)>max1)max1=length($1);if(length($2)>max2)max2=length($2)}END{for(i=1;i<=max2;i+=width){printf("%"(max1+1)"s","");for(pos=i;pos<i+width&&pos<=max2;pos+=10)printf(pos-i>width-10?"%s":"%-10s",pos);print"";for(j in seq){printf("%"max1"s ",name[j]);for(k=i;k<=max2&&k<i+width;k++)printf("%s","\033[38;2;0;0;0m\033[48;2;"a[seq[j][k]]"m"seq[j][k]"\033[0m");print""}}}' "width=${1-60}" <(aacolor|sed $'s/ /\t/;s/ /;/g') -)
# print an alignment of selected viruses
thin 92 <spike.pid|cut -d\ -f1|seqkit grep -f- spike.aa|seqkit grep -nrvp 'SARS coronavirus'|cat - <(seqkit grep -nrip db159,hu-1,tor2,zc45,rp3\\b,rs7896,banal-20-52,ratg,lyra11,btsy2,shc014,wiv1\\b,gx_p2v,gx-p1e spike.aa)|seqkit rmdup -s|aln --reorder -|cut -d, -f1|sed 's/>[^ ]*/>/;s/ ORF.*//;s/ surface .*//;s/Severe acute respiratory syndrome/SARS/;s/Khosta-2 strain //;s/ isolate / /;s/ strain / /;s/Bat SARS-like coronavirus //;s/Betacoronavirus sp. //;s/ RNA$//'|seqcol
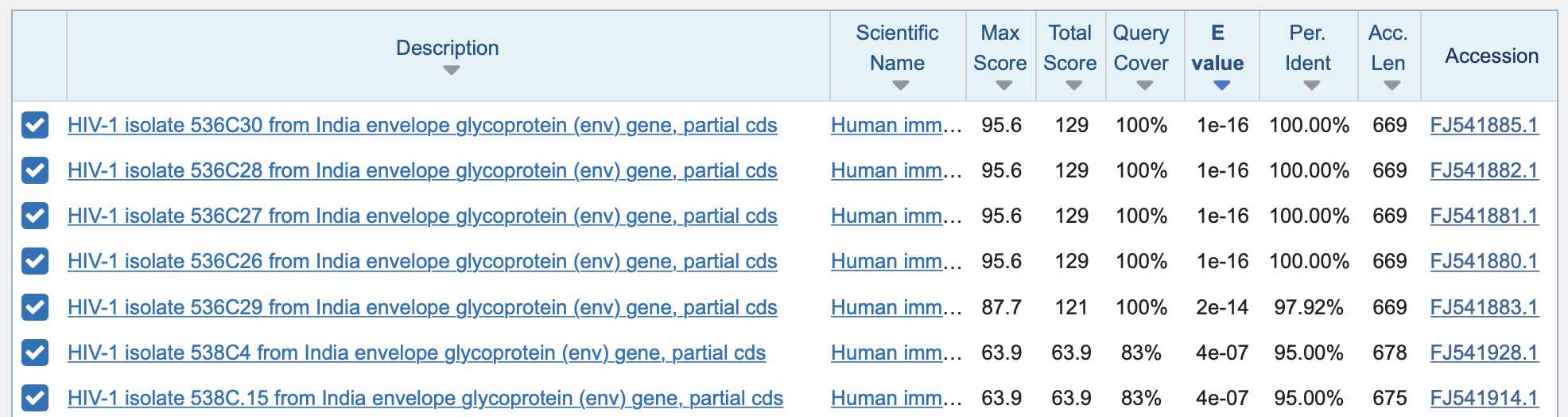
Arkmedic did a BLAST search for the nucleotide sequence of Pradhan's first insert, but he used the coding sequence of the insert in SARS2 and not HIV. He restricted the search to viruses published in 2019 or earlier, so many of the top matches were sequences of HIV: [https://www.arkmedic.info/p/absolute-proof-the-gp-120-sequences]
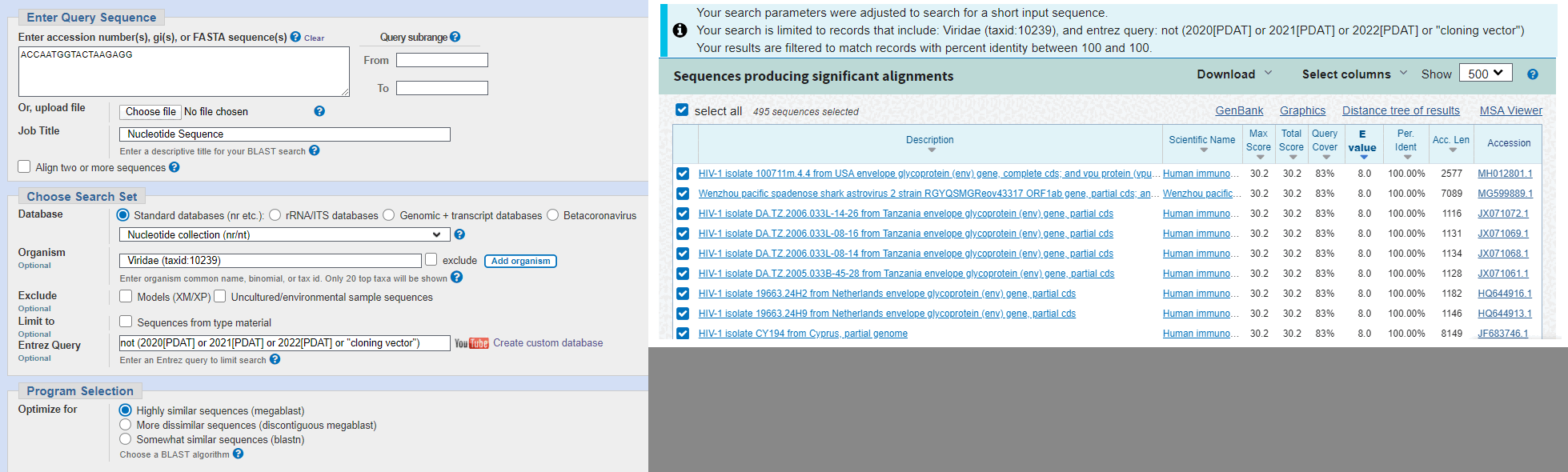
However his top matches got only 83% query coverage because they only matched 15 out of 18 bases of the query, similar to my matches here:
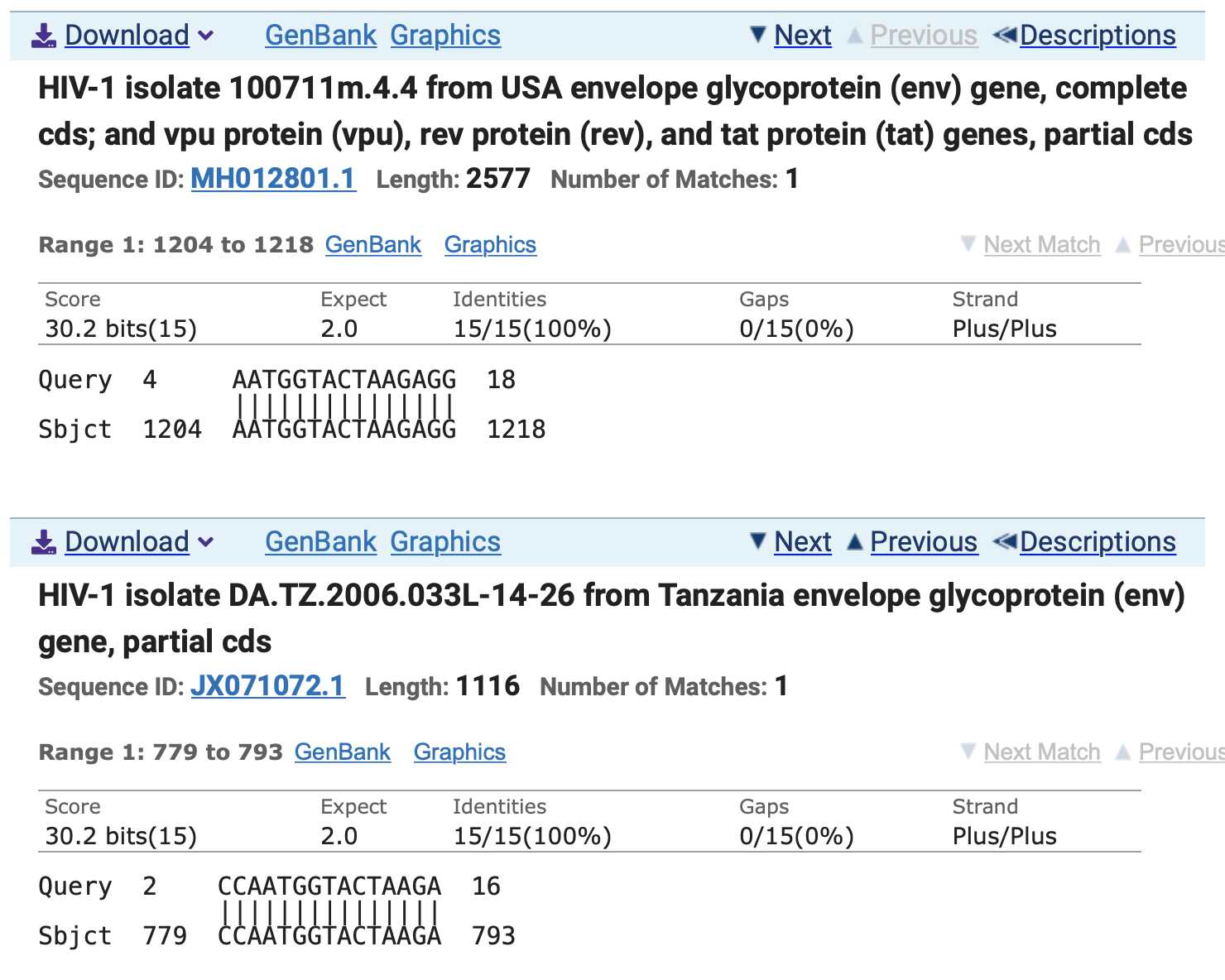
The first match above was missing the first 3 bases of the query and
the second match was missing the first base, but both matches were still
in frame so that the first match coded for 5 out of 6 aa of
TNGTKR and the second match coded for 4 out of 6 aa:
$ emu()(curl -sd "id=$(paste -sd,)" "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=${1:-nuccore}&rettype=${2:-fasta}&retmode=$3")
$ emu protein fasta_cds_na<<<AVN55366|seqkit translate|seqkit subseq -r 400:409
>lcl|MH012801.1_cds_AVN55366.1_1 [gene=env] [protein=envelope glycoprotein] [protein_id=AVN55366.1] [location=1..2577] [gbkey=CDS]
WENGTKRSNE
$ emu '' fasta_cds_na<<<JX071072|seqkit translate|seqkit subseq -r 259:266
>lcl|JX071072.1_cds_AFV81607.1_1 [gene=env] [protein=envelope glycoprotein] [protein_id=AFV81607.1] [location=<1..>1116] [gbkey=CDS]
NPNGTKIY
The main text of Pradhan's paper doesn't include the accession numbers of the HIV sequences which matched their inserts, but you have to dig them up from the supplementary Excel file which was only posted at bioRxiv but not ResearchGate: https://www.biorxiv.org/content/10.1101/2020.01.30.927871v1.supplementary-material?versioned=true. It shows that the first insert was matched by 10 different sequences from Thailand:
| country | accession | subtype | insert |
|---|---|---|---|
| Thailand | AFU28737.1 | CRF01_AE | Insert 1 |
| Thailand | AFU28711.1 | CRF01_AE | Insert 1 |
| Thailand | AFU28717.1 | CRF01_AE | Insert 1 |
| Thailand | AFU28733.1 | CRF01_AE | Insert 1 |
| Thailand | AFU28693.1 | CRF01_AE | Insert 1 |
| Thailand | AFU28721.1 | CRF01_AE | Insert 1 |
| Thailand | AFU28699.1 | CRF01_AE | Insert 1 |
| Thailand | AFU28729.1 | CRF01_AE | Insert 1 |
| Thailand | AFU28705.1 | CRF01_AE | Insert 1 |
| Thailand | AFU28725.1 | CRF01_AE | Insert 1 |
| Kenya | ALB06757.1 | G | Insert 2 |
| India | ACL98861.1 | C | Insert 3 |
| India | ACL98864.1 | C | Insert 3 |
| India | ACL98860.1 | C | Insert 3 |
| India | ACL98859.1 | C | Insert 3 |
| India | AKR75206.1 | C | Insert 4 |
However in all Thai sequences the first insert was coded by
ACA AAT GGA ACC AAG AGG, so it's actually 3 bases different
from the nucleotide sequence in Wuhan-Hu-1 that was used by Arkmedic
(ACC AAT GGT ACT AAG AGG):
$ emu()(curl -sd "id=$(paste -sd,)" "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=${1:-nuccore}&rettype=${2:-fasta}&retmode=$3")
$ emu protein fasta_cds_na<<<AFU28737,AFU28711,AFU28717,AFU28733,AFU28693,AFU28721,AFU28699,AFU28729,AFU28705,AFU28725 >insert1.fa
$ seqkit translate insert1.fa|seqkit locate -p TNGTKR|sed 1d|cut -f1,5|while read l m;do seqkit grep -p $l insert1.fa|seqkit subseq -r $[m*3-2]:$[m*3-2+17];done|seqkit seq -s|sort -u|sed 's/.../& /g'
ACA AAT GGA ACC AAG AGG
There were about 3.2 million results at GenBank when I searched for
0:2019[dp] viruses[organism], but about 0.9 million or about
27% of all results were classified under HIV-1.
[https://www.ncbi.nlm.nih.gov/nuccore/?term=0%3A2019%5Bdp%5D+viruses%5Borganism%5D]
So the reason why many of Arkmedic's top hits at BLAST were sequences of
HIV-1 might be because before 2020 HIV-1 was probably the species of
virus with the most sequences at GenBank:
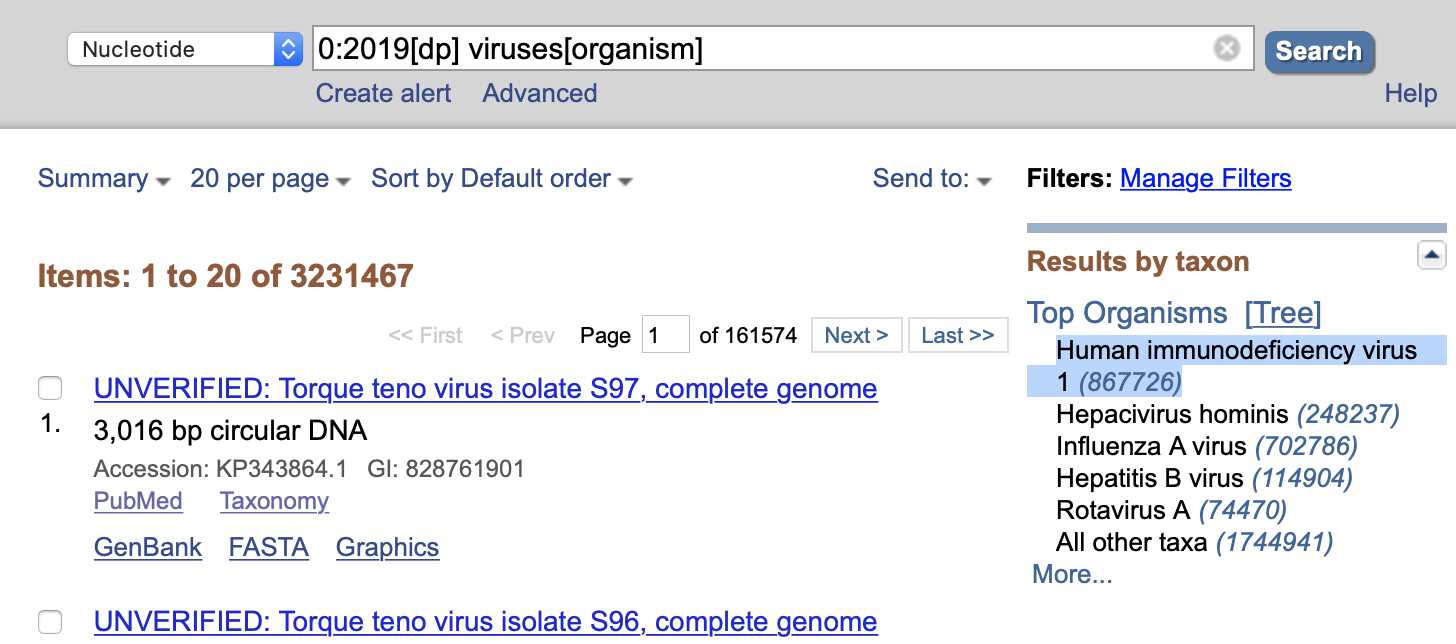
Arkmedic also wrote that he didn't find any 100% match when he
searched for the nucleotide sequence of Pradhan's second insert on
BLAST, but that's because he again used the nucleotide sequence in
Wuhan-Hu-1 (CAC AAA AAC AAC AAA AGT), which is 3 bases
different from Pradhan's HIV sequences:
$ emu protein fasta_cds_na<<<ALB06757 >insert2.fa $ x=$(seqkit translate insert2.fa|seqkit locate -p HKNNKS|sed 1d|cut -f5);seqkit subseq -r $[x*3-2]:$[(x+5)*3] insert2.fa|seqkit seq -s|sed 's/.../& /g' CAT AAA AAT AAT AAA AGT
When I did a BLAST search for the nucleotide sequence in HIV, it had 18/18 matching bases only with Pradhan's Kenyan HIV-1 sequence and with some other random virus where the match was on the minus strand (but in the screenshot below the other results with 100% identity and 100% query coverage were noncontiguous matches where the match was split into two different segments, as you can see from the max score being lower than on the first two rows):
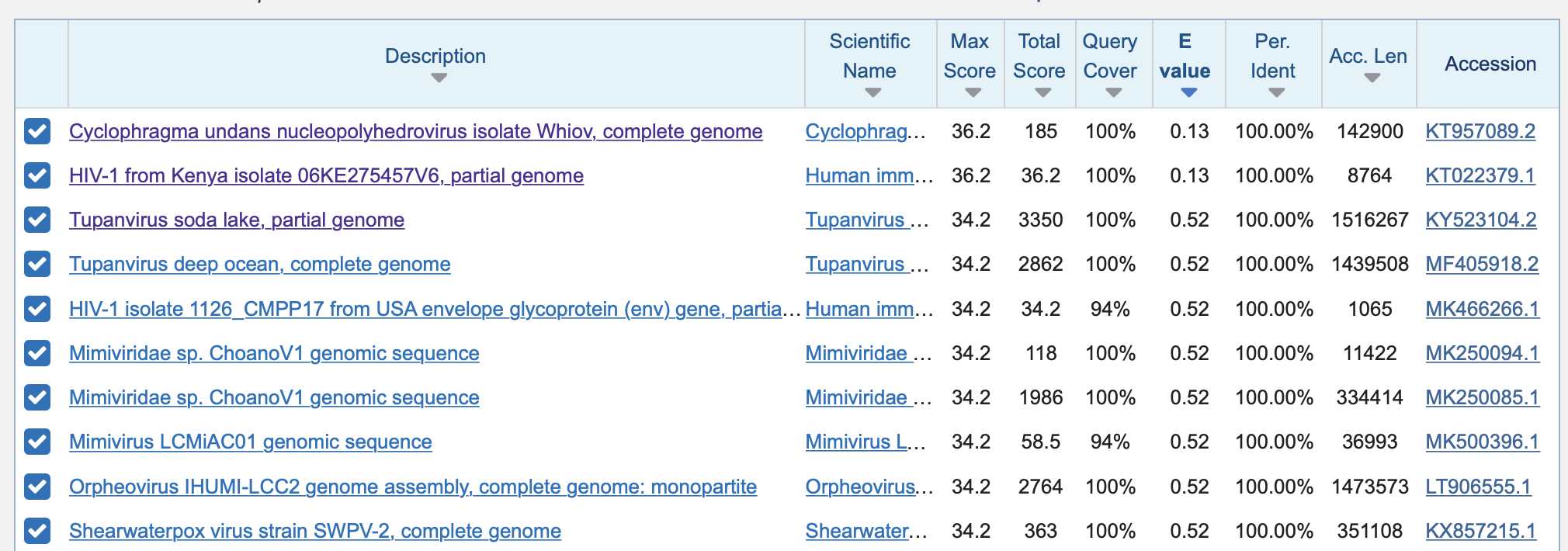
Arkmedic also pointed out that he found no matches for Pradhan's
third insert on BLAST, but that's because he searched for the region
around the insert in SARS-CoV-2 and not HIV (RSYLTPGDSSSG
and not RTYLFNETRGNSSSG). The nucleotide sequence of the
version in HIV is found in all 4 HIV sequences that were listed in
Pradhan's supplementary Excel file:
$ emu protein fasta_cds_na<<<ACL98861,ACL98864,ACL98860,ACL98859 >insert3.fa
$ x=RTYLFNETRGNSSSG;seqkit translate insert3.fa|seqkit locate -p $x|sed 1d|cut -f1,5|while read l m;do seqkit grep -p $l insert3.fa|seqkit subseq -r $[m*3-2]:$[(m+${#x})*3];done|seqkit seq -s|sort -u|sed 's/.../& /g'
AGG ACA TAC CTG TTT AAT GAG ACA AGA GGT AAT TCA AGC TCA GGT AAT
Arkmedic wrote: "TLDR: In order to get the 3 inserts of Gp-120 to exist in SARS-Cov-2, the genomic sequences that coded for them had to have got there by recombination from another organism or in a lab. Because they don't exist anywhere in nature it is not possible to have come from another organism."
However as I showed earlier, Pradhan's first insert actually looks like a deletion in SARS1 and not an insertion in SARS2. And Pradhan's second and third inserts were placed at the wrong spot because he only did a pairwise alignment of SARS1 and SARS2 and he didn't include more sarbecoviruses in his alignment.
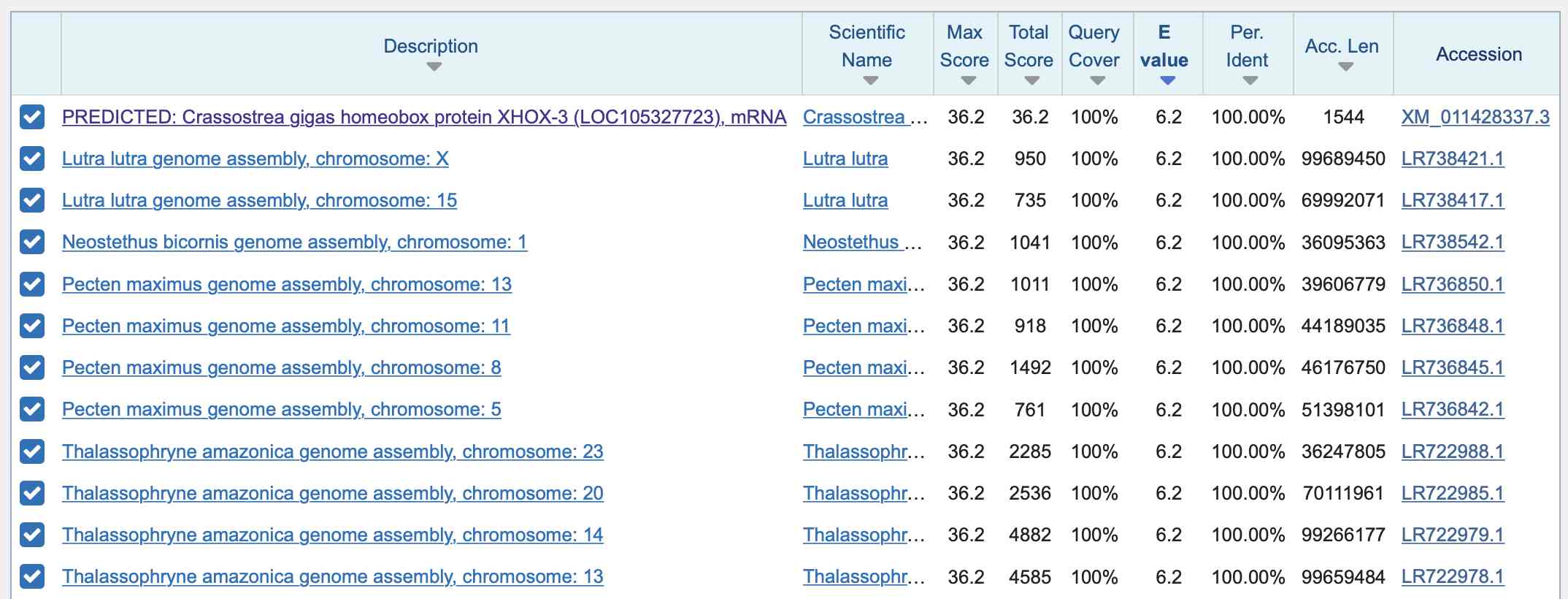
Arkmedic was also wrong that the inserts "don't exist anywhere in nature", because the first two so-called inserts are only 6 aa long, so if you do a BLAST search for the nucleotide sequences of the inserts in Wuhan-Hu-1, they get perfect matches to many organisms but just not to virus sequences published before 2020, like for example this shows that the second insert has perfect matches to mollusks, otters, fish, and so on:
Arkmedic wrote: "I mean, sure, the old virus can
undergo some mutations (GATTTCA...) but these are evolutionarily very slow
and can result in deletions and changes. In order to get inserts for
these to happen by chance is so rare that it would take millions of
years to develop functional inserts by chance so you really need a
genome donor." However for example the nucleotide sequence of the
second insert is CACAAAAACAACAAAAGT in Wuhan-Hu-1, which is
identical in RaTG13, differs by one base in BANAL-52 and BtSY2, and
differs by two bases in Rp22DB159. And even if you consider all
sequences released since 2020 to be fake, even in ZC45 which was
published in 2018, the region of the so-called insert differs by 3
nucleotide changes and 3 gaps:
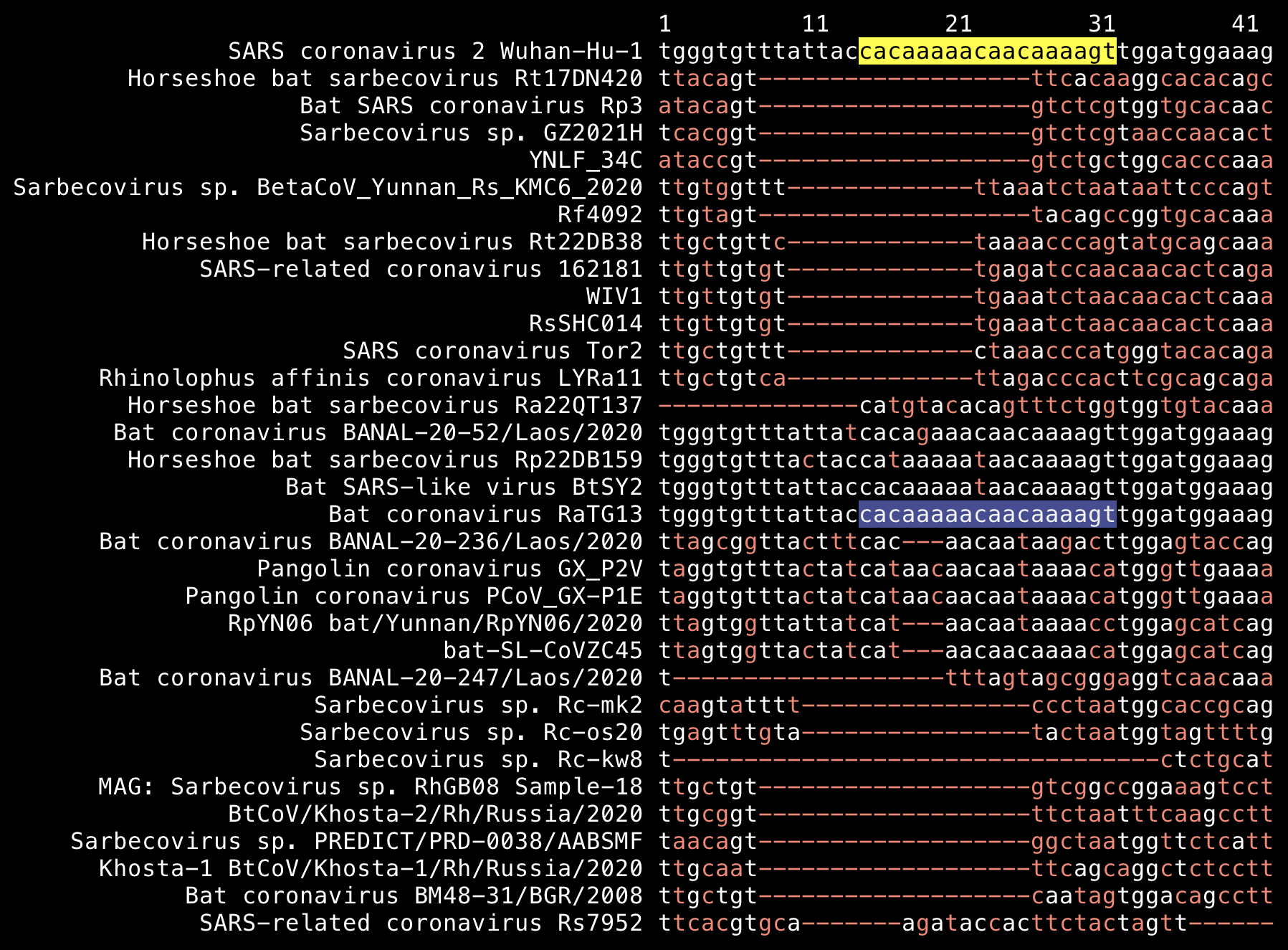
Similarly the nucleotide sequence of the first insert TNGTKR differs by 2 bases in RaTG13, 3 bases in BANAL-52, BtSY2, and Rp22DB159, and 7 bases in ZC45 (but it's located within a variable region of the genome that is expected to be mutating fast):
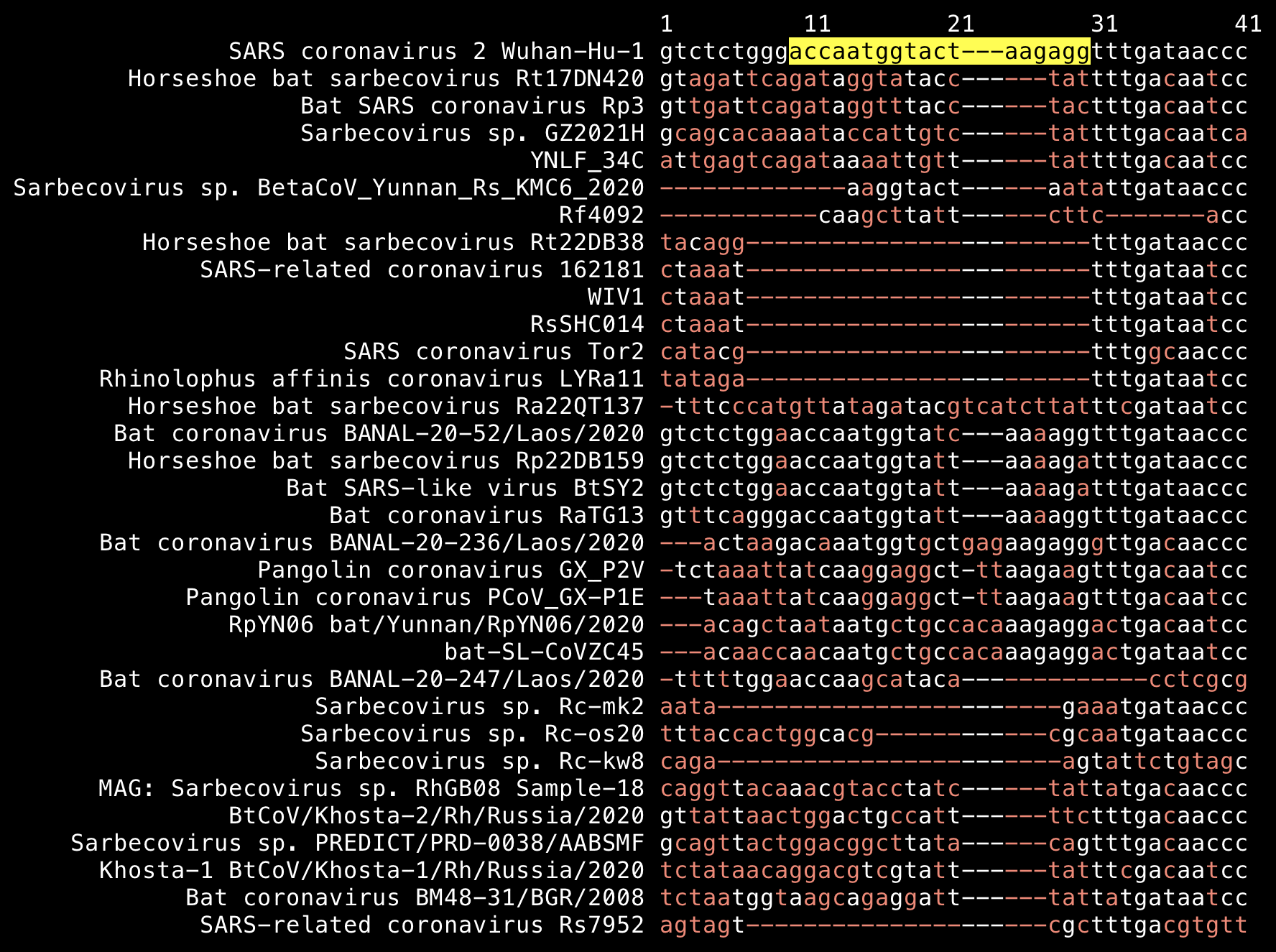
From the HIV sequence database of the Los Alamos National Laboratory, you can download a FASTA file which contains 2-5 sequences from different subtypes of HIV-1 along with 27 chimpanzee and gorilla sequences of SIV that are similar to HIV-1. Set "Alignment type" to "Web (all complete sequences)", set "DNA/Protein" to "Protein", and click "Get alignment": https://www.hiv.lanl.gov/content/sequence/NEWALIGN/align.html. You can keep region as "env" to download only the envelope protein, which covers about 20% of the genome of HIV-1.
The 2023 edition of LANL's subtype reference has a total of 555
sequences, but zero of them have a perfect match to Pradhan's first
motif TNGTKR. Two sequences have one mismatch, 215 have 2
mismatches, and the rest have 3 mismatches:
$ wget sars2.net/f/HIV1_REF_2023_env_DNA.fasta
$ x=TNGTKR;for((i=0;i<=${#x};i++));do seqkit seq -g HIV1_REF_2023_env_PRO.fasta|seqkit replace -sp \# -r X|seqkit locate -iPp $x -m$i|sed 1d|awk '!a[$1]++'|echo "$i `wc -l`";done
0 0
1 2
2 215
3 555
4 555
5 555
6 555
For the second motif HKNNKS, there's zero perfect
matches and only sequence with a single mismatch:
$ x=HKNNKS;for((i=0;i<=${#x};i++));do seqkit seq -g HIV1_REF_2023_env_PRO.fasta|seqkit replace -sp \# -r X|seqkit locate -iPp $x -m$i|sed 1d|awk '!a[$1]++'|echo "$i `wc -l`";done
0 0
1 1
2 41
3 551
4 555
5 555
6 555
All of the matches with one mismatch were located in the first half of gp120:
$ seqkit seq -g HIV1_REF_2023_env_PRO.fasta|seqkit replace -sp\# -rX|seqkit locate -ip TNGTKR -m1|cut -f1,3,5-|column -t seqID pattern start end matched Ref.H.GB.00.00GBAC4001.FJ711703 tngtkr 190 195 tngtnr Ref.L.CD.01.L_CG_0018a_01.MN271384 tngtkr 132 137 tngttr $ seqkit seq -g HIV1_REF_2023_env_PRO.fasta|seqkit replace -sp\# -rX|seqkit locate -ip HKNNKS -m1|cut -f1,3,5-|column -t seqID pattern start end matched Ref.101_01B.CN.13.YNKM250.MK158946 hknnks 366 371 hfnnks
The two matches to TNGTKR were both located within
variable loops but the match to HKNNKS wasn't:
seqkit grep -nrp FJ711703,MN271384,YNKM250,HXB2 HIV1_REF_2023_env_PRO.fasta|seqkit replace -p'^Ref\.(.*)\..*?$' -r\$1|mafft --quiet -|seqkit fx2tab|gawk -F\\t 'ARGIND==1{a[$1]=$2}ARGIND==2{for(i=$2;i<=$3;i++)loop[i]=$1}ARGIND==3{if($1~/HXB2/){gap=0;split($2,z,"");for(i in z){if(z[i]=="-")gap++;gaps[i]=gap}}name[FNR]=$1;split($2,z,"");for(i in z)seq[FNR][i]=z[i];if(length($1)>max1)max1=length($1);if(length($2)>max2)max2=length($2)}END{width=60;for(i=1;i<=max2;i+=width){print"";printf("%*s",max1+1,"");for(pos=i;pos<i+width&&pos<=max2;pos+=10)printf(pos-i>width-10?"%s":"%-10s",pos-gaps[pos]);print"";printf("%*s",max1+1,"");for(pos=i;pos<i+width;pos++){l=loop[pos-gaps[pos]];printf"%s",l?l:" "};print"";for(j in seq){printf("%"max1"s ",name[j]);for(k=i;k<=max2&&k<i+width;k++)printf("%s","\033[38;2;0;0;0m\033[48;2;"a[seq[j][k]in a?seq[j][k]:"X"]"m"seq[j][k]"\033[0m");print""}}}' <(aacolor|sed $'s/ /\t/;s/ /;/g') <(printf %s\\n 1:131:157 2:158:196 3:296:331 4:385:418 5:460:469|tr : \\t) -
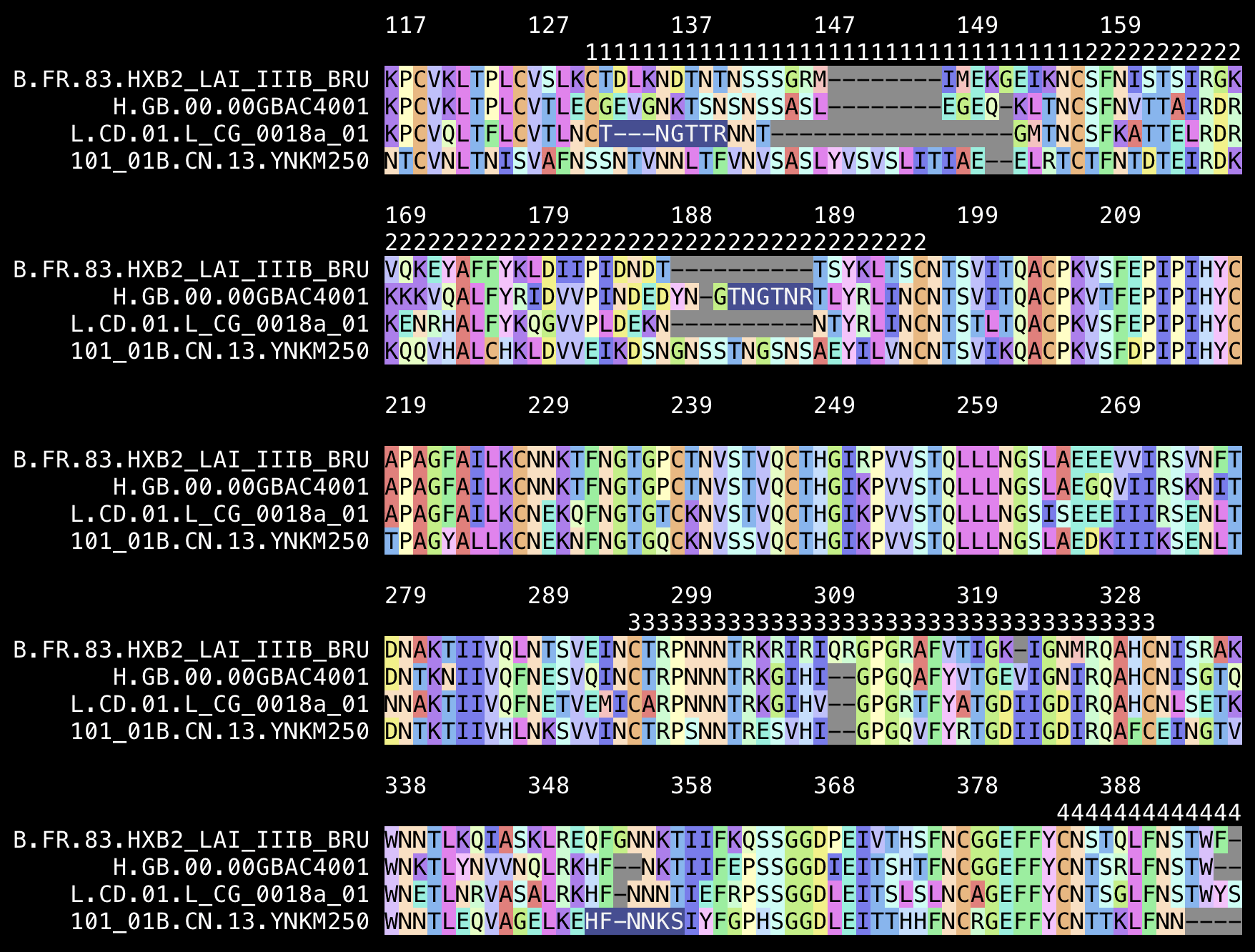
The numbers in the image above show the position in HXB2. I used these as the coordinates of the variable loops in the env protein of HXB2:
loop start end V1 131 157 V2 158 196 V3 296 331 V4 385 418 V5 460 469
The E value or expect value indicates how many similarly close matches are expected to occur by chance for the given query sequence and pool of searched sequences.
You can try to go to the web GUI for blastp:
https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp.
Then set query to TNGTKR, organism to "Viruses (taxid:10239)", click "Add organism", set the second organism to "SARS-CoV-2 (taxid:2697049)" and click the exclude
checkbox next to it. It showed me that the matches to HIV got an E value
of about 372, which means that about 372 similar matches with 6 out of 6
identical residues are expected to occur by chance. There were a total
of 76 perfect matches with 6 out of 6 identical residues, so the expect
score was about 5 times higher than the actual number of perfect
matches.
When I did another search in the whole nr database so that I didn't include the organism filters, then the E scores of perfect matches increased to about 54093. So they were now about 145 times higher than with the filters because now the pool of subject sequences was much larger, and the E score is directly proportional to the effective length of the pool of subject sequences.
The E score is calculated by the formula
E = m * n * 2^(-S), where m and n
are the effective length of the query and database sequences and
S is the bitscore. I don't know how to calculate the
effective length, because it is calculated by subtracting a parameter
called the length adjustment from the actual length to account for
duplicate sequences and repetitive elements within sequences, so I
believe you need to download the entire database to calculate the length
adjustment. However since the E values are directly proportional to the
effective length of the pool of subject sequences, from the ratio of my
E scores you can calculate that the effective length of the whole nr
database is about 145 times higher than the effective length of the
subset of the nr database that contains viruses apart from
SARS-CoV-2.
Pradhan's second insert HKNNKS was 6 residues long like
the first insert, but when I searched for the second insert among
viruses other than SARS-CoV-2 in the nr database, the E value was now
about 185 for the match to HIV. The results included 36 perfect matches,
so the expect score was again about 5 times higher than the number of
perfect matches. I believe the reason why both queries don't get the
same E value even though they're the same length is that the algorithm
assigns different scores to common amino acids and rare amino acids.
The nr database contains a massive number of protein sequences for a
few species of widely studied human viruses like SARS-CoV-2, HIV-1, and
influenza A. But in order to restrict the search to a database that only
has one or a few representative samples for each species of virus, you
can search in the refseq_protein database instead and restrict the
organism to viruses. It gave me an E value of about 33 for results that
had a perfect match to Pradhan's first insert TNGTKR (but
the results didn't include any HIV sequences, because the envelope
protein of the HIV-1 refseq matches only 2 of 6 residues of
TNGTKR at the spot where Pradhan's HIV sequences from
Thailand match TNGTKR):
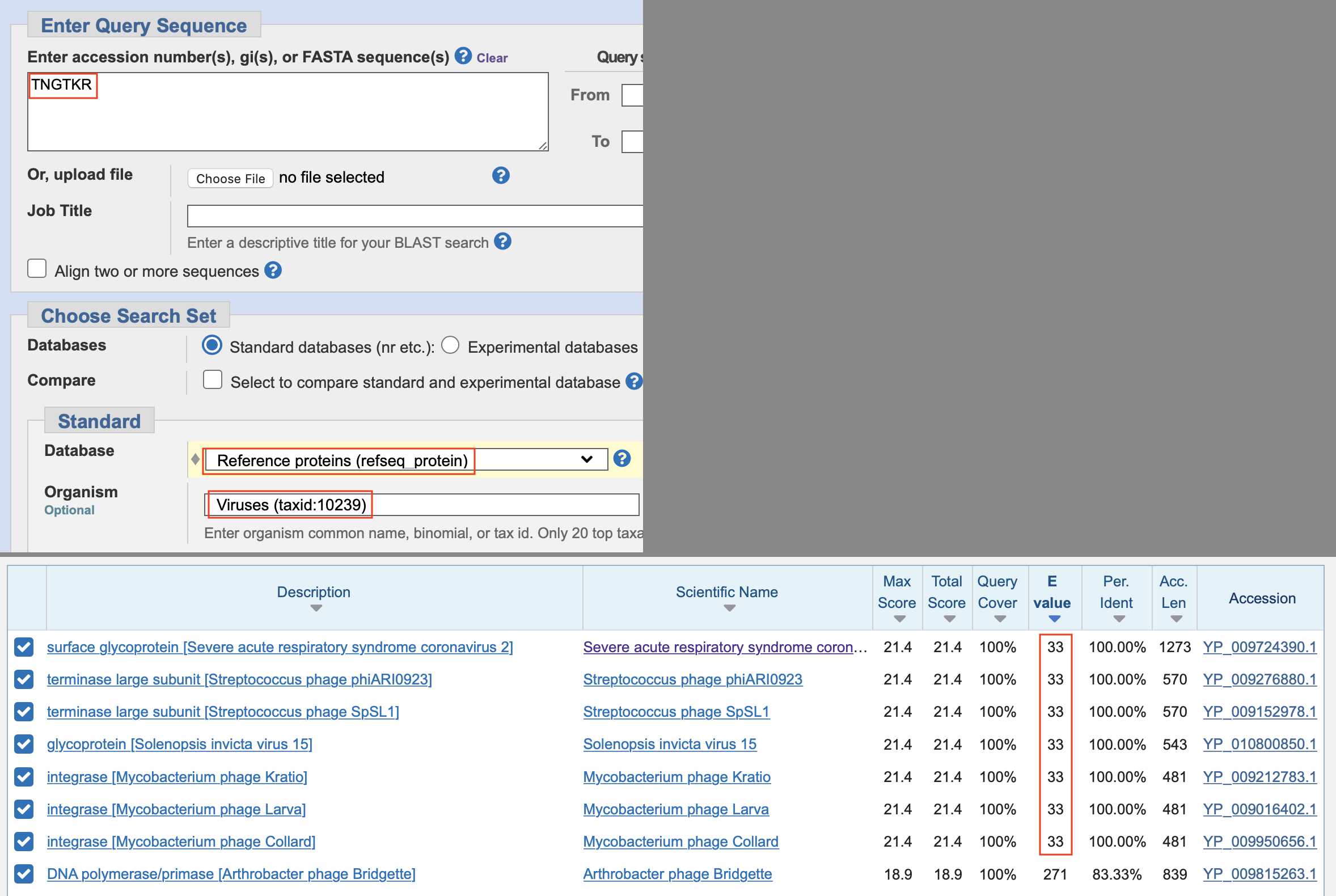
So even within a fairly small pool of proteins from virus refseqs, the E values for perfect matches to the 6 aa insert were still very high.
In the following code I created my own BLAST database for proteins
from virus refseqs. For some reason it failed to download coding
sequences for about half of all virus refseqs even though refseqs should
usually have annotations for proteins. But anyway, I now got only a
single perfect match to TNGTKR which was in the refseq for
SARS-CoV-2, but the E value of the match was still 289 which is very
high:
$ wget -q https://ftp.ncbi.nlm.nih.gov/refseq/release/viral/viral.1.1.genomic.fna.gz $ seqkit seq -ni viral.fa|curl -sd "id=$(paste -sd, -)" 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta_cds_na'>viral.aa $ makeblastdb -dbtype prot -in viral.aa [...] $ echo $'>a\nTNGTKR'|blastp -db viral.aa -evalue 1000 [...] >lcl|NC_045512.2_prot_YP_009724390.1_3 [gene=S] [locus_tag=GU280_gp02] # spike protein of refseq for SARS-CoV-2 [db_xref=GeneID:43740568] [protein=surface glycoprotein] [protein_id=YP_009724390.1] [location=21563..25384] [gbkey=CDS] Length=1273
Score = 16.5 bits (31), Expect = 289, Method: Composition-based stats.
Identities = 6/6 (100%), Positives = 6/6 (100%), Gaps = 0/6 (0%)
Query 1 TNGTKR 6
TNGTKR
Sbjct 73 TNGTKR 78
[...]
Tony VanDongen posted this tweet: [https://x.com/tony_vandongen/status/1887707580148941305]
Brian Gadd posted this reply: "AFAIK there is no evidence that these sequences are necessary or sufficient for CD4 binding; they are absent from most gp120 sequences, no two homology sequences are found in the same molecule of gp120, they barely overlap with the CD4 binding site & not at the most important contact points." [https://x.com/Brian_Gadd/status/1887724003604746578] He also replied: "Brunetti et al. did look at spike-CD4 interaction and their binding data suggests that this interaction is almost exclusively mediated by the RBD, not the NTD: https://elifesciences.org/articles/84790." Another user responded: "CD209 (DC-SIGN) is a lectin that binds HIV gp120 and modulates CD4+ T-cell infection. Lectin-gp120 interactions impact HIV entry—critical for viral transmission and immune evasion." But Gadd answered: "Yes. And Amraei et al. found that L-SIGN and DC-SIGN do indeed bind to spike, and that this interaction is mediated by the RBD: https://pubs.acs.org/doi/10.1021/acscentsci.0c01537."
In April 2020 Jean-Claude Perez and Luc Montagnier published a preprint at OSF titled "COVID-19, SARS and Bats Coronaviruses Genomes Unexpected Exogenous RNA Sequences". [https://osf.io/preprints/osf/d9e5g_v1] A slightly revised version of the preprint was published in May 2020 at ResearchGate. [https://www.researchgate.net/publication/341756383_COVID-19_SARS_and_Bats_Coronaviruses_Genomes_Unexpected_Exogenous_RNA_Sequences] The paper was published in August 2020 in an Indian junk journal. [https://www.granthaalayahpublication.org/journals/granthaalayah/article/view/IJRG20_B07_3568] I refer to the author of the paper as simply Perez, who seems to have done most of the work on the paper.
Perez confusingly did not use Wuhan-Hu-1 as his reference genome but another early sequence from Wuhan. [https://www.ncbi.nlm.nih.gov/nucleotide/LR757998] It's missing the first 25 bases from the start of Wuhan-Hu-1, so the genome coordinates given by Perez are off by 25 relative to Wuhan-Hu-1 coordinates.
Perez identified two contiguous regions with a high number of matches to HIV or SIV, which he called region A and region B:

Perez indicated that region A was 600 bases long but it matched bases 21072 to 21672 of the reference genome, which would mean that region A would be 601 bases long if both ends of the range would be inclusive. But the end of the range is exclusive and Perez started numbering the bases from zero and not one, so the region matches bases 21073 to 21672 in regular one-based numbering where both ends of the range are inclusive:
$ emu()(curl -sd "id=$(paste -sd,)" "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=${1:-nuccore}&rettype=${2:-fasta}&retmode=$3")
$ emu<<<LR757998>perezref.fa
$ seqkit locate -p AGGGTTTTTTCACTTACATTTGTGGGTTTATACAACAAAAGCTAGCTCTTGGAGGTTCCGTGGCTATAAAGATAACAGAACATTCTTGGAATGCTGATCTTTATAAGCTCATGGGACACTTCGCATGGTGGACAGCCTTTGTTACTAATGTGAATGCGTCATCATCTGAAGCATTTTTAATTGGATGTAATTATCTTGGCAAACCACGCGAACAAATAGATGGTTATGTCATGCATGCAAATTACATATTTTGGAGGAATACAAATCCAATTCAGTTGTCTTCCTATTCTTTATTTGACATGAGTAAATTTCCCCTTAAATTAAGGGGTACTGCTGTTATGTCTTTAAAAGAAGGTCAAATCAATGATATGATTTTATCTCTTCTTAGTAAAGGTAGACTTATAATTAGAGAAAACAACAGAGTTGTTATTTCTAGTGATGTTCTTGTTAACAACTAAACGAACAATGTTTGTTTTTCTTGTTTTATTGCCACTAGTCTCTAGTCAGTGTGTTAATCTTACAACCAGAACTCAATTACCCCCTGCATACACTAATTCTTTCACACGTGGTGTTTATTACCCTGACAAAGTTTTCAGATCC perezref.fa|cut -f5,6|column -t
start end
21073 21672
Regions A and B combined consist of bases 21098 to 22027 of Wuhan-Hu-1. Region A consists of the last 458 bases of ORF1ab, the 7-base intergenic region between ORF1ab and the spike protein, and the first 135 bases of the spike protein. Region B consists of bases 136 to 465 of the spike protein.
Table 1 of Perez's paper showed matches to regions A and B in various sequences of HIV-1, HIV-2, and SIV, where matches shorter than 18 bases were indicated as not significant (even though in reality all matches were either so short or they had so many errors that their E values were far above 0.05).
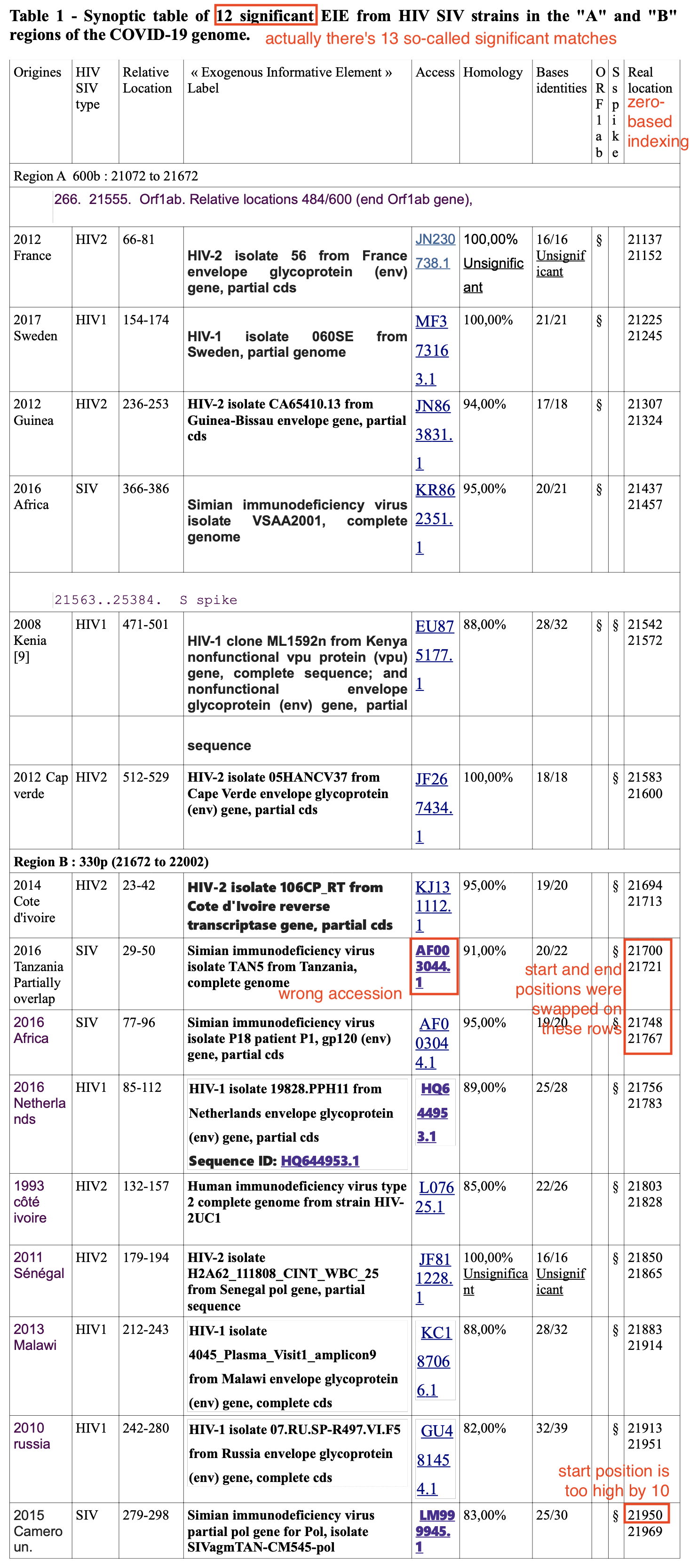
The table has several errors:
It's also confusing that the starting and ending positions in the last column of the table use zero-based indexing and not regular one-based indexing.
When I downloaded all 15 sequences in the table and I did a BLAST search for the sequences against SARS-CoV-2, there were no matches returned when I ran BLAST with the default settings:
$ cat perez # fixed errors mentioned above and converted to 1-based indexing JN230738.1 21138 21153 MF373163.1 21226 21246 JN863831.1 21308 21325 KR862351.1 21438 21458 EU875177.1 21543 21573 JF267434.1 21584 21601 KJ131112.1 21695 21714 AF003044.1 21749 21768 # the starting and ending positions on these JN091691.1 21701 21722 # two rows were accidentally swapped HQ644953.1 21757 21784 L07625.1 21804 21829 JF811228.1 21851 21866 KC187066.1 21884 21915 GU481454.1 21914 21952 LM999945.1 21941 21970 # the starting position of this row was too high by 10 $ curl "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=$(cut -d\ -f1 perez|paste -sd, -)">perez.fa $ brew install blast [...] $ makeblastdb -dbtype nucl -in perez.fa [...] $ fmt='sseqid evalue nident length pident sstrand qstart qend' $ <perezref.fa blastn -db perez.fa -outfmt "6 $fmt" $ # no results
So therefore in order to use less strict matching criteria, I added
the options -word_size 5 -evalue 100 which gave me one or
more matches for all 15 sequences. At first the starting position of one
of my matches wasn't the same as in Perez's table, but I got the
starting positions to match when I added the option
-task blastn:
$ <perezref.fa blastn -db perez.fa -outfmt "6 $fmt" -task blastn|awk '$7>=21073&&$8<=22027'|awk 'NR==FNR{a[$1]=$2" "$3;pos[$1]=NR;next}{print pos[$1],$0,a[$1]}' perez -|awk '{print$0,$8==$10&&$9==$11?"true":"false"}'|sort -rk12|sort -sgk3|awk '!a[$1]++'|sort -n|cut -d\ -f2-|(echo $fmt perezstart perezend matchesperez;cat)|column -t
sseqid evalue nident length pident sstrand qstart qend perezstart perezend matchesperez
JN230738.1 1.3 16 16 100.000 plus 21138 21153 21138 21153 true
MF373163.1 0.003 21 21 100.000 minus 21226 21246 21226 21246 true
JN863831.1 4.7 17 18 94.444 minus 21308 21325 21308 21325 true
KR862351.1 0.11 20 21 95.238 minus 21438 21458 21438 21458 true
EU875177.1 0.009 28 32 87.500 minus 21543 21573 21543 21573 true
JF267434.1 0.11 18 18 100.000 plus 21584 21601 21584 21601 true
KJ131112.1 0.38 19 20 95.000 minus 21695 21714 21695 21714 true
AF003044.1 0.38 19 20 95.000 plus 21749 21768 21749 21768 true
JN091691.1 0.38 20 22 90.909 minus 21701 21722 21701 21722 true
HQ644953.1 0.009 25 28 89.286 plus 21757 21784 21757 21784 true
L07625.1 1.3 22 26 84.615 plus 21804 21829 21804 21829 true
JF811228.1 1.3 16 16 100.000 minus 21851 21866 21851 21866 true
KC187066.1 0.009 28 32 87.500 minus 21884 21915 21884 21915 true
GU481454.1 0.009 32 39 82.051 minus 21914 21952 21914 21952 true
LM999945.1 0.38 25 30 83.333 minus 21941 21970 21941 21970 true
In the table above the E value indicates how many matches are expected to occur by chance for a given query in a given database. A bigger database size means higher E values. But even though in the code above I had a tiny database that consisted of only 15 sequences of HIV and SIV, 4 out of 15 matches above got an E value above 1, which means that more than 1 similar matches are expected to occur by chance. And even the lowest E value was only about 0.003.
The output above shows that 10 out of 15 matches are on the minus strand, which means that HIV/SIV matched the reverse complement of the segment in SARS-CoV-2, so the matched segment does not even code for the same amino acids in SARS-CoV-2 and HIV/SIV. For example the second sequence matches a Swedish HIV sequence from 2017, but it matches SARS-CoV-2 on the opposite strand so the translation is completely different: [https://www.ncbi.nlm.nih.gov/nuccore/MF373163]
SARS-CoV-2: HIV:
<--------------------
--------------------> TACGAAGTCTACTACTGCGTA # reversed version of segment matched in SARS-CoV-2
ATGCGTCATCATCTGAAGCAT ATGCTTCAGATGATGACGCAT # reversed version with complemented bases
AATGCGTCATCATCTGAAGCATTT TATGCTTCAGATGATGACGCATAT
N A S S S E A F Y A S D D D A Y
So after the 10 matches on the wrong strand are eliminated, only 5 out of 15 matches remain. But 2 out of the 5 matches are in a different frame in SARS-CoV-2 and HIV/SIV, so their amino acid translation is not the same, which means that only 3 out of the 15 matches in Perez's table are both on the right strand and in the right frame:
| accession | SC2 start | SC2 CDS start | SC2 frame | HIV start | HIV CDS start | HIV frame | same frame |
|---|---|---|---|---|---|---|---|
| JN230738 | 21138 | 20899 | 1 | 77 | 77 | 2 | false |
| JF267434 | 21584 | 47 | 2 | 284 | 284 | 2 | true |
| AF003044 | 21749 | 212 | 2 | 845 | 845 | 2 | true |
| HQ644953 | 21757 | 220 | 1 | 967 | 967 | 1 | true |
| L07625 | 21804 | 267 | 3 | 6701 | 23 | 2 | false |
In the table above the "CDS start" columns
show the starting position of the match within the protein coding
sequence it is part of. For example the match on the first row starts at
base 20899 of ORF1ab in SARS-CoV-2. By calculating
(20899-1)%3+1, you get the frame number which is 1.
In order to see how high the E values of Perez's matches were in the full nucleotide database, I copied this sequence which consists of Perez's regions A and B:
$ seqkit subseq -r 21073:22027 sars2.fa|seqkit seq -s GTTACAAAAGAAAATGACTCTAAAGAGGGTTTTTTCACTTACATTTGTGGGTTTATACAACAAAAGCTAGCTCTTGGAGGTTCCGTGGCTATAAAGATAACAGAACATTCTTGGAATGCTGATCTTTATAAGCTCATGGGACACTTCGCATGGTGGACAGCCTTTGTTACTAATGTGAATGCGTCATCATCTGAAGCATTTTTAATTGGATGTAATTATCTTGGCAAACCACGCGAACAAATAGATGGTTATGTCATGCATGCAAATTACATATTTTGGAGGAATACAAATCCAATTCAGTTGTCTTCCTATTCTTTATTTGACATGAGTAAATTTCCCCTTAAATTAAGGGGTACTGCTGTTATGTCTTTAAAAGAAGGTCAAATCAATGATATGATTTTATCTCTTCTTAGTAAAGGTAGACTTATAATTAGAGAAAACAACAGAGTTGTTATTTCTAGTGATGTTCTTGTTAACAACTAAACGAACAATGTTTGTTTTTCTTGTTTTATTGCCACTAGTCTCTAGTCAGTGTGTTAATCTTACAACCAGAACTCAATTACCCCCTGCATACACTAATTCTTTCACACGTGGTGTTTATTACCCTGACAAAGTTTTCAGATCCTCAGTTTTACATTCAACTCAGGACTTGTTCTTACCTTTCTTTTCCAATGTTACTTGGTTCCATGCTATACATGTCTCTGGGACCAATGGTACTAAGAGGTTTGATAACCCTGTCCTACCATTTAATGATGGTGTTTATTTTGCTTCCACTGAGAAGTCTAACATAATAAGAGGCTGGATTTTTGGTACTACTTTAGATTCGAAGACCCAGTCCCTACTTATTGTTAATAACGCTACTAATGTTGTTATTAAAGTCTGTGAATTTCAATTTTGTAATGATCCATTTTTGGGTGTTTATTACCACAAAAACAACAAAAGTTGGATGGAAAGT
Then I pasted the sequence to the text box here: https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn. I changed the database type to "Nucleotide collection (nt/nr)", which used to be the default database back when Perez wrote his paper, and which has slightly more HIV sequences than the new core_nt database. I entered "HIV-1 (taxid:11676)" in the organism field, clicked "Add Organism", entered "HIV-2 (taxid:11709)", clicked "Add Organism", and entered "Simian immunodeficiency virus (taxid:11723)". I typed "0:2019[dp]" under "Entrez Query" to only select sequences with date of publication before 2020. And then under "Program selection" change the algorithm from "megablast" to "blastn", because otherwise short matches would not be returned. And under "Algorithm parameters" I increased "Expect threshold" from 0.05 to 10000, so that even matches that have more than 5% likelihood to be due to chance will be returned, and I also increased "Max target sequences" from 100 to 5000. Then I clicked "BLAST".
There was a single match which had an E value of about 1.4, which was the Swedish HIV sequence that was shown on the second row of Perez's Table 1, and which has 21 out of 21 matching bases. But the next best match had an E value of about 5.0, which means that about 5.0 similarly close matches were expected to occur by chance:
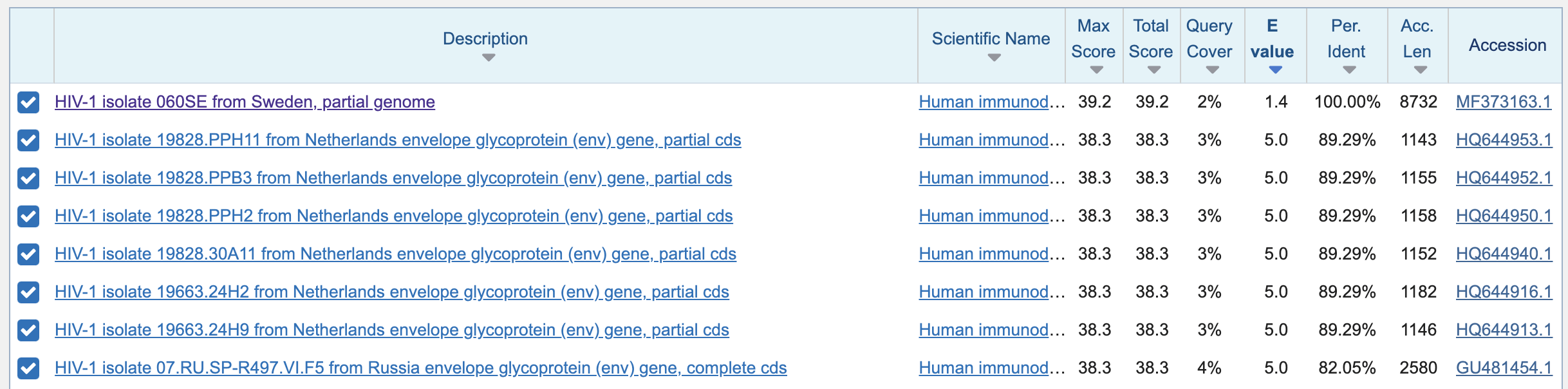
If I would've omitted the step where I increased the expect threshold from 0.05 to 1000, BLAST would have returned zero results and said "No significant similarity found", because none of the matches had lower than 5% likelihood of occuring by chance.
Next I selected "Hit Table (text)" from the "Download" menu and kept only results which were included in Perez's Table 1. The expect value was over 100 for 9 out of 15 results:
| subject | expect | pident | length | mismatch | qstart | qend | sstart | send | sstrand |
|---|---|---|---|---|---|---|---|---|---|
| JN230738.1 HIV-2 isolate 56 from France envelope glycoprotein (env) gene, partial cds | 746 | 100.000 | 16 | 0 | 91 | 106 | 77 | 92 | + |
| MF373163.1 HIV-1 isolate 060SE from Sweden, partial genome | 1.4 | 100.000 | 21 | 0 | 179 | 199 | 5634 | 5614 | - |
| JN863831.1 HIV-2 isolate CA65410.13 from Guinea-Bissau envelope gene, partial cds | 2604 | 94.444 | 18 | 1 | 261 | 278 | 436 | 419 | - |
| KR862351.1 Simian immunodeficiency virus isolate VSAA2001, complete genome | 61 | 95.238 | 21 | 1 | 391 | 411 | 8635 | 8615 | - |
| EU875177.1 HIV-1 clone ML1592n from Kenya nonfunctional vpu protein (vpu) gene, complete sequence; and nonfunctional envelope glycoprotein (env) gene, partial sequence | 5.0 | 87.500 | 32 | 4 | 496 | 526 | 270 | 239 | - |
| JF267434.1 HIV-2 isolate 05HANCV37 from Cape Verde envelope glycoprotein (env) gene, partial cds | 61 | 100.000 | 18 | 0 | 537 | 554 | 284 | 301 | + |
| KJ131112.1 HIV-2 isolate 106CP_RT from Cote d'Ivoire reverse transcriptase gene, partial cds | 214 | 95.000 | 20 | 1 | 648 | 667 | 85 | 66 | - |
| AF003044.1 Simian immunodeficiency virus isolate P18 patient P1, gp120 (env) gene, partial cds | 214 | 95.000 | 20 | 1 | 702 | 721 | 845 | 864 | + |
| JN091691.1 Simian immunodeficiency virus isolate TAN5 from Tanzania, complete genome | 214 | 90.909 | 22 | 2 | 654 | 675 | 2801 | 2780 | - |
| HQ644953.1 HIV-1 isolate 19828.PPH11 from Netherlands envelope glycoprotein (env) gene, partial cds | 5.0 | 89.286 | 28 | 3 | 710 | 737 | 967 | 994 | + |
| L07625.1 Human immunodeficiency virus type 2 complete genome from strain HIV-2UC1 | 746 | 84.615 | 26 | 4 | 757 | 782 | 6701 | 6726 | + |
| JF811228.1 HIV-2 isolate H2A62_111808_CINT_WBC_25 from Senegal pol gene, partial sequence | 746 | 100.000 | 16 | 0 | 804 | 819 | 803 | 788 | - |
| KC187066.1 HIV-1 isolate 4045_Plasma_Visit1_amplicon9 from Malawi envelope glycoprotein (env) gene, complete cds | 2604 | 90.000 | 20 | 2 | 849 | 868 | 431 | 412 | - |
| GU481454.1 HIV-1 isolate 07.RU.SP-R497.VI.F5 from Russia envelope glycoprotein (env) gene, complete cds | 5.0 | 82.051 | 39 | 5 | 867 | 905 | 1064 | 1029 | - |
| LM999945.1 Simian immunodeficiency virus partial pol gene for Pol, isolate SIVagmTAN-CM545-pol | 214 | 83.333 | 30 | 5 | 894 | 923 | 1098 | 1069 | - |
Perez and other people have posted the image below on Twitter. [https://x.com/JCPEREZCODEX/status/1920908991925776671] But the image misleadingly doesn't show the expect value or strand of the matches, so I added the green and red pieces of text to the image which show the expect value and strand. The matches with the right strand and frame are shown in green, and matches with the wrong strand or frame are shown in red. There's only two green matches, but both of them have a very high E-value.
I took the E-values from the table in the previous section, which shows the E-values when I searched for regions A and B of Wuhan-Hu-1 against sequences in the nt database with the species HIV-1, HIV-2, or SIV. Regions A and B account for about one fiftieth part of the length of the whole genome of SARS-CoV-2, so if I would've searched for the whole genome instead of only regions A and B, then the E-values would've been about 50 times bigger.
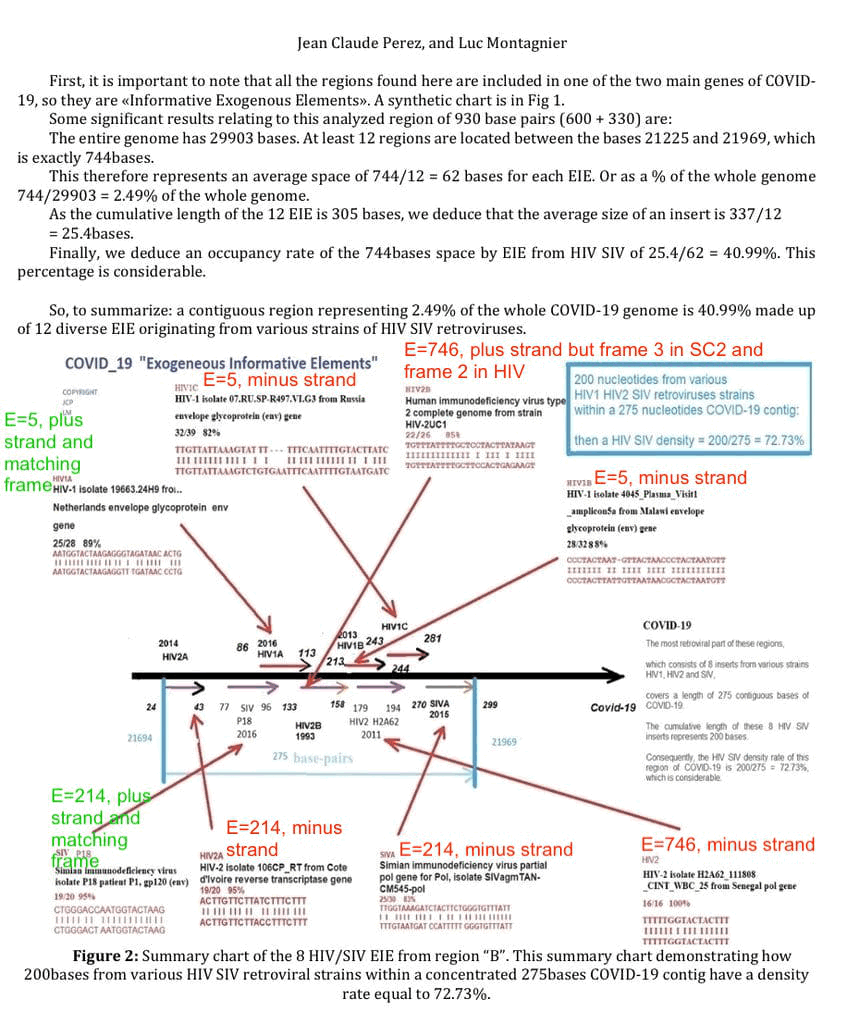
In Perez's graphic it's also misleading that he drew the arrows of all 8 matches from left to right, even though 5 out of 8 matches were on the minus strand so the arrows should've been drawn from right to left.
In Table 2 of the paper by Perez and Montagnier, there's a list of 4 additional matches to HIV and SIV that are not located in the regions A or B:
However 3 out of 4 matches in the table are on the minus strand:
$ curl 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=LR757998'>perezref.fa
$ printf %s\\n 'KM378564.1 8752 8770' 'EU184986.1 14341 14378' 'AY516986.1 20374 20401' 'HQ217329.1 20401 20430'>perez2
$ cut -d\ -f1 perez2|curl -sd "id=$(paste -sd,)" 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta'>perez2.fa
$ makeblastdb -dbtype nucl -in perez2.fa
[...]
$ fmt='sseqid evalue nident length pident sstrand sstart send qstart qend'
$ <perezref.fa blastn -db perez2.fa -outfmt "6 $fmt" -task blastn|awk 'NR==FNR{a[$1]=$2" "$3;pos[$1]=NR;next}{print pos[$1],$0,a[$1]}' perez2 -|awk '{print$0,$8==$10&&$9==$11?"true":"false"}'|sort -rk12|awk '!a[$1]++'|sort -n|cut -d\ -f2-|(echo $fmt perezstart perezend matchesperez;cat)|column -t
sseqid evalue nident length pident sstrand qstart qend perezstart perezend matchesperez
KM378564.1 3.8 22 27 81.481 minus 4696 4722 8752 8770 false
EU184986.1 4.95e-05 33 38 86.842 minus 14341 14378 14341 14378 true
AY516986.1 4.95e-05 26 28 92.857 plus 20374 20401 20374 20401 true
HQ217329.1 4.95e-05 28 30 93.333 minus 20401 20430 20401 20430 true
The first row of Perez's table is shown to match positions 8751 to 8770 of his reference genome (i.e. positions 8752 to 8770 using one-based indexing and an inclusive end of the range). But that region of his reference genome does not actually match the HIV sequence on the first row even though the region 8736 to 8755 shown in my output above does, so Perez may have accidentally entered the wrong starting and ending positions on the first row.
Only one out of the 4 matches in Table 2 is on the plus strand, but it doesn't even match HIV, because it matches a part of a human genome that is included in an HIV integration site sequence, where the first 69 bases come from an HIV long terminal repeat but the rest of the sequence comes from a human genome: [https://www.ncbi.nlm.nih.gov/nuccore/AY516986]
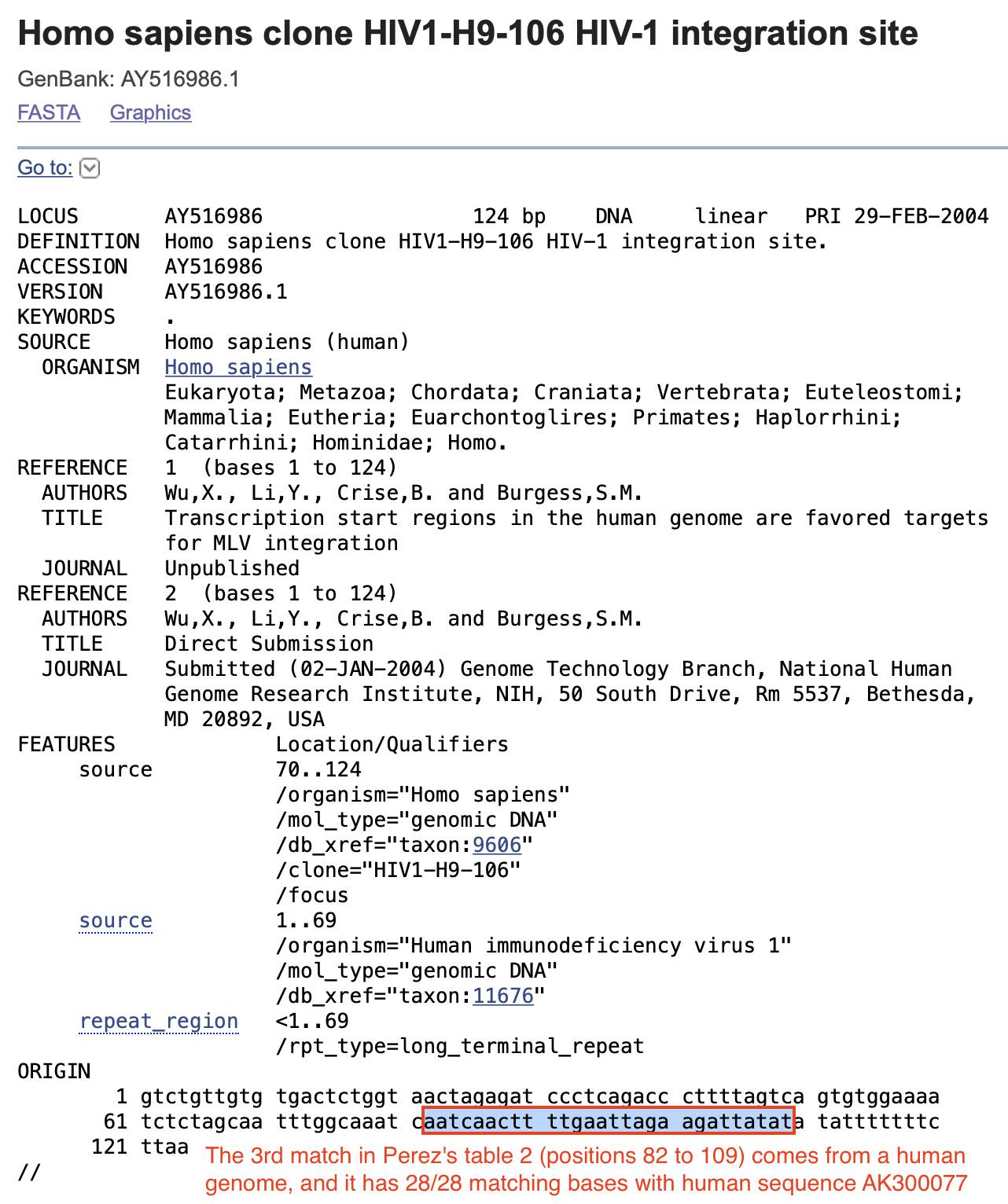
Perez wrote that in the match to the integration site sequence "addresses 20373 to 20401 comes from an HIV1 Integrase from a USA virus from 2004". However he may have confused the term "integration site" with the HIV integrase enzyme which is contained within the pol gene. The integrase is located at positions 4236 to 5086 in HXB2, but the first 69 bases of the integration site sequence are identical to bases 112 to 180 of HXB2 (which is contained within the 5' long terminal repeat):
$ emu<<<NC_001802>hiv1.fa $ emu<<<AY516986|seqkit subseq -r1:69|seqkit locate -if- hiv1.fa|cut -f4-7|column -t strand start end matched + 112 180 gtctgttgtgtgactctggtaactagagatccctcagacccttttagtcagtgtggaaaatctctagca
Another big fail by Perez was that he thought that one of his BLAST matches was located at the start of the spike protein in RaTG13 but that it spanned both ORF1ab and the spike protein in SARS-CoV-2. But that's because his nonstandard reference genome was missing the first 25 bases of Wuhan-Hu-1, so he should've subtracted 25 from Wuhan-Hu-1 coordinates when he identified the start of the spike protein:

On page 36 of the paper Perez presented this match to the rodent parasite Plasmodium yoelii:

However the matching sequence is in the frame 2 in SARS-CoV-2 but frame 1 in Plasmodium yoelii, so Perez's amino acid translation of the match in SARS-CoV-2 is wrong:
$ url='https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta_cds_na' $ curl "$url&id=MN908947">sars2.na $ curl "$url&id=LM993664">yoelii10.na $ seqkit locate -pCACAAGTCAAACAAATTTACAAAACACCACCAATTAAAGAT sars2.na|cut -f4-6|column -t strand start end + 2348 2388 # (2348-1)%3+1 is frame 2 $ seqkit locate -pCACAAATCAAACAAATTTACAAAACACAAACCAAAAAAAAAT yoelii10.na|cut -f4-6|column -t strand start end + 106 147 # (106-1)%3+1 is frame 1 + 106 147
Next Perez presented a second match to the same rodent parasite which they initially said appeared to be located downstream in the same protein as the first match, even though actually the match was located in a different chromosome and a different protein, and in fact Perez later corrected his own initial impression and wrote that "it is therefore clear that this second region of Yoelii does not coincide with the extension downstream of the sequence 'Fam a'". However for some reason Perez still decided to concatenate the two unrelated matches together. The second match to Plasmodium yoelii has two gaps near the start in SARS-CoV-2 which means that the segments before and after the gaps are in different frames:

The screenshot above shows that the match to the whole chromosome in Plasmodium yoelii is on the minus strand, but the match to the gene in Plasmodium yoelii is still on the plus strand because the gene is on the reverse strand. And the part of the match after the gaps happens to be in the +2 frame in both Plasmodium yoelii and SARS-CoV-2. But the match is still not impressive at all because it has only 36 out of 46 identical bases so the percentage identity is only about 78%:
$ url='https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta_cds_na' $ curl "$url&id=MN908947">sars2.na $ curl "$url&id=LK934642">yoelii14.na # protein coding sequences within chromosome 14 $ seqkit locate -pCAAAACACCACCAATTAAAGATTTTGGTGGTTTTAATTTTTCACAA sars2.na|cut -f4-6|column -t strand start end + 2367 2412 # (2367-1)%3+1 is frame 3 $ seqkit locate -pCAAAATAAAACCAATTATATATTTTGATCATATTAATTTTTCAAAA yoelii14.na|cut -f4-6|column -t strand start end + 6903 6948 # (6903-1)%3+1 is frame 3
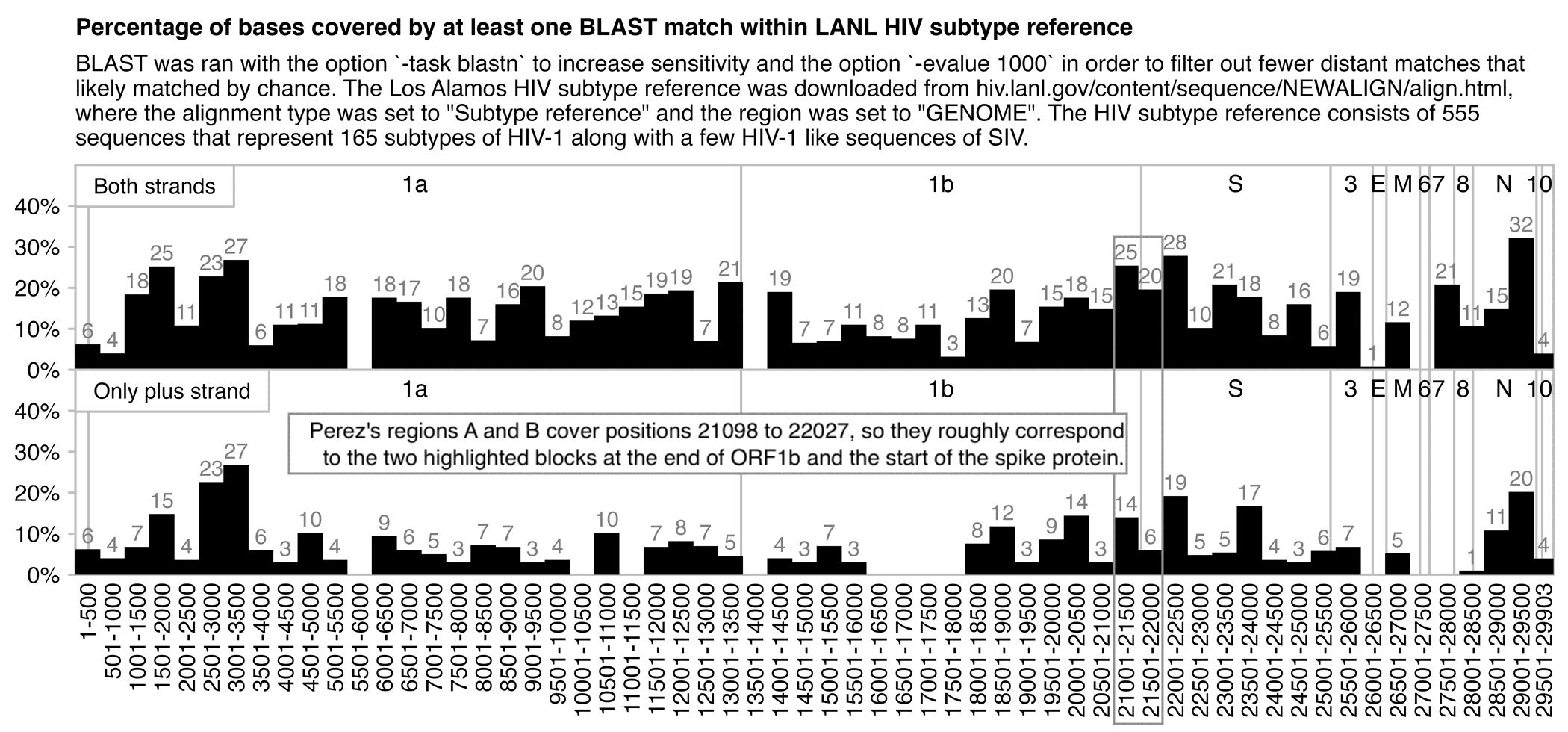
In the plot below I used web BLAST to search for Wuhan-Hu-1 within the nr/nt database, where I set species to HIV, task to blastn, expect threshold to 1000, and maximum results to 1000. I then kept scrolling the alignment view for a couple of minutes to fetch all results, I saved the source code of the web page, and I made a list of the starting and ending positions of each match, and I counted how many positions were covered by at least one match. The E values of the matches ranged between 5.2 and 776. When I divided Wuhan-Hu-1 to 500-base blocks, the three blocks with the highest covered percentage were the blocks that started at positions 21001, 11001, and 21501, where two of the blocks roughly coincide to Perez's regions A and B which cover positions 21098 to 22027. However there was also a block in ORF3a which had almost as high coverage percentage as the block at the end of ORF1b that covered the first half of regions A and B. And when I included only matches on the plus strand, the block at the end of ORF1b got 0% coverage, and the block at the start of the spike protein got 14% coverage which was about as high as three random blocks in ORF1ab:
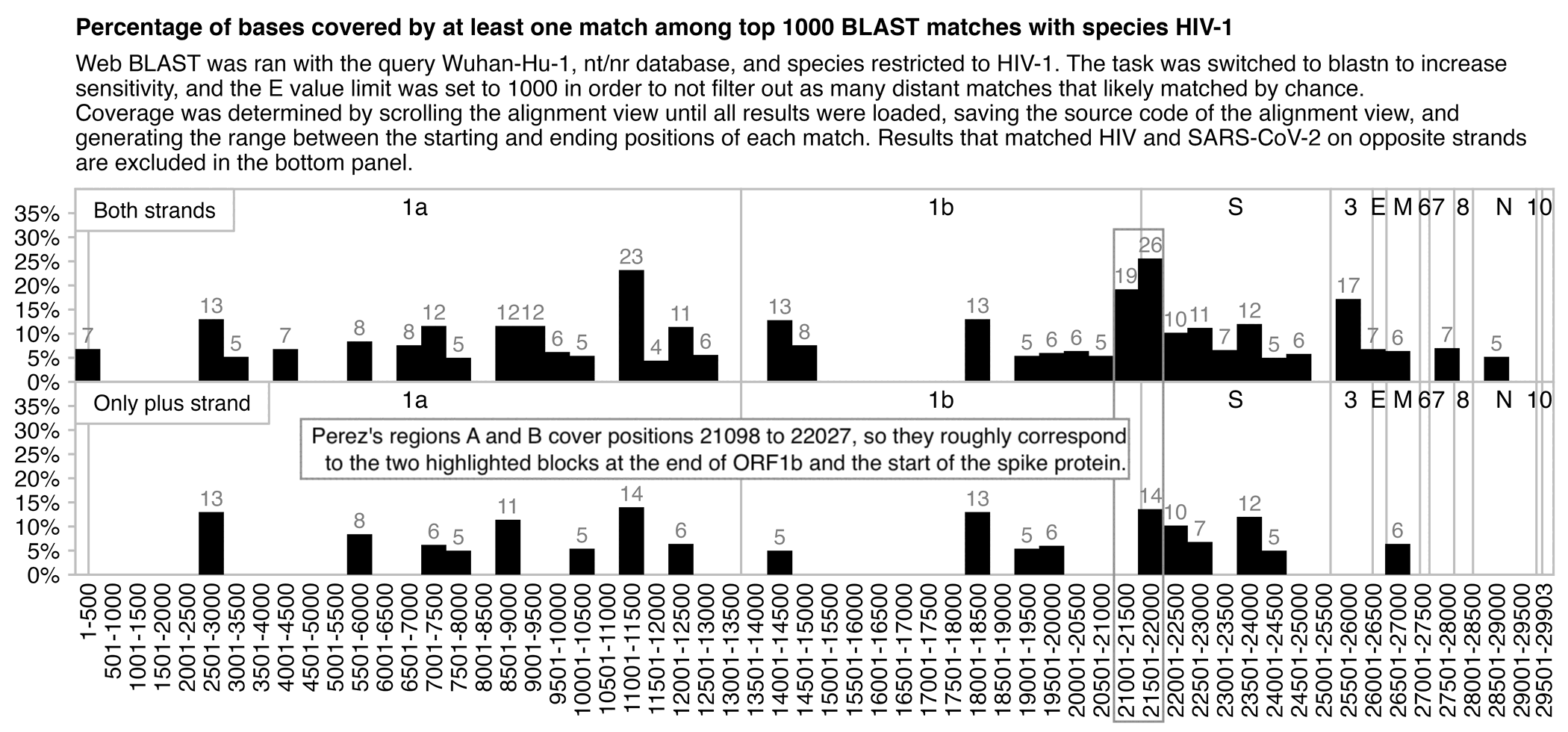
Even though I included the top 1000 BLAST results in the plot above,
there were many duplicate matches where different sequences of HIV
matched the same segment, so there were only 82 unique starting
positions among the matches. But next in order to search within a
diverse set of HIV subtypes with only a few results by subtype, I did a
BLAST search within the HIV subtype reference published by the Los
Alamos National Laboratory:
https://www.hiv.lanl.gov/content/sequence/NEWALIGN/align.html.
It contains 555 sequences that represent 165 different subtypes of
HIV-1, but it also contains a few HIV-1 like SIV sequences from chimps
and gorillas. I got zero results when I searched for Wuhan-Hu-1 against
the LANL subtype reference with the default options, but my number of
results increased to 7 when I added the option -task blastn
and to 609 when I also added the option -evalue 1000. There
were 245 unique starting positions among the results, so now the matches
were distributed more evenly because there weren't as many duplicate
matches for the same region. And now even in the top panel which
includes both strands, the two bars which roughly correspond to Perez's
regions A and B only had the 4th and 11th highest percentage of covered
bases:
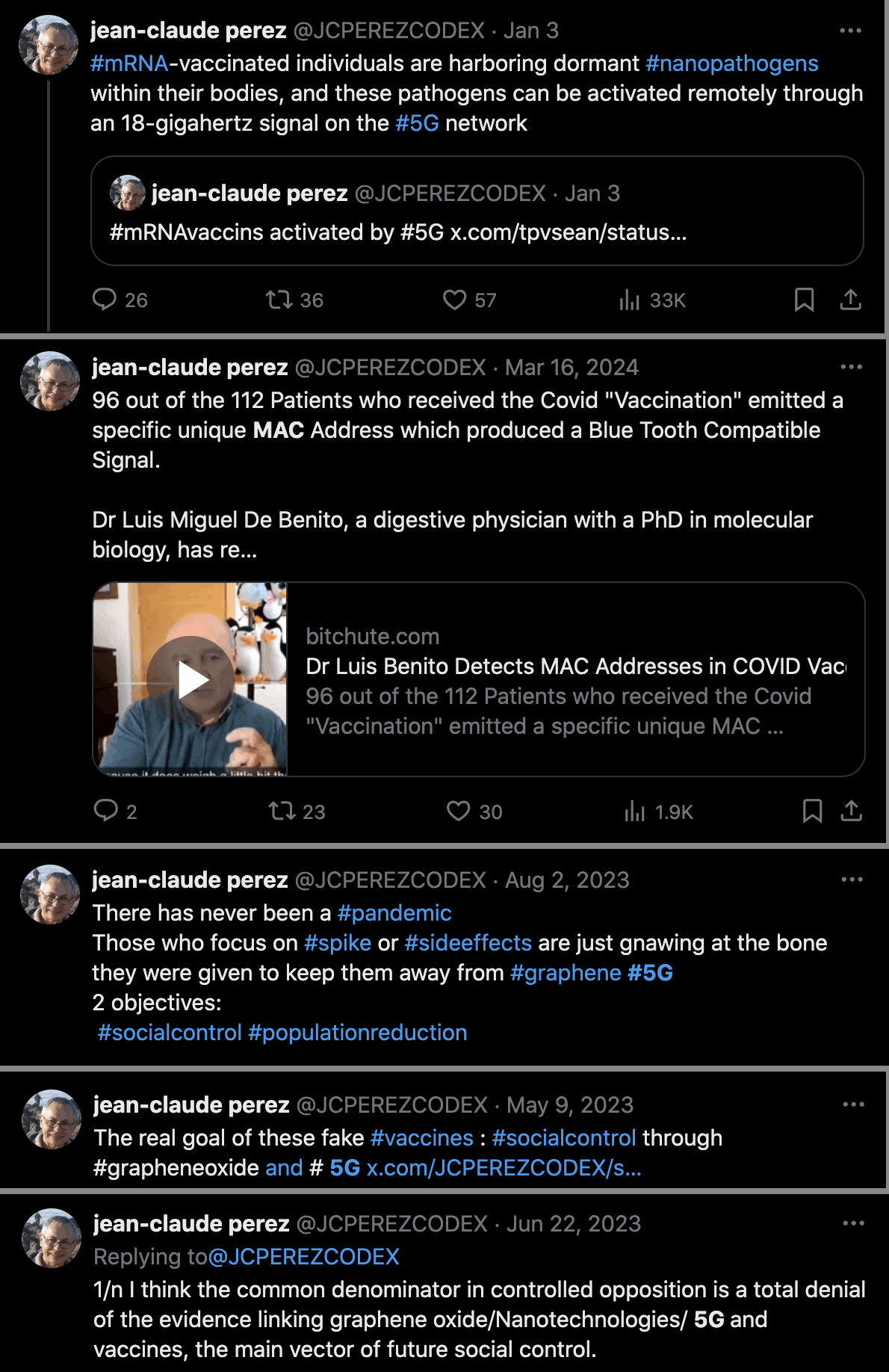
Perez might have written his paper as a form of deliberate disinformation, because he has also said that vaccinated people are harboring nanopathogens that can be activated by 5G, that vaccinated people have MAC addresses, that vaccines contain graphene oxide, and that the pandemic was fake: [https://x.com/JCPEREZCODEX/status/1875105196294201653, https://x.com/JCPEREZCODEX/status/1768971965178548510, https://x.com/JCPEREZCODEX/status/1686762168606167040, https://x.com/JCPEREZCODEX/status/1655898773774647303, https://x.com/JCPEREZCODEX/status/1671902292402995200]