This HTML file addresses misconceptions about the reference genome of SARS-CoV-2 that have emerged among people who claim that viruses do not exist.
In the paper by Wu et al. where the Wuhan-Hu-1 reference genome was described, the authors wrote that when they did metagenomic sequencing for a sample of lung fluid from a COVID patient, they got a total of 384,096 contigs when they did de-novo assembly for the reads using MEGAHIT, and they used the longest contig as the initial reference genome of SARS2. Some no-virus people thought it meant that the other contigs were alternative candidates for the genome of SARS2, but actually the other contigs mostly consisted of fragments of the human genome or bacterial genomes. In Supplementary Table 2 which shows the best match on BLAST for the 50 most abundant contigs, the best match is bacterial for all other contigs except the contig for SARS2.
Wu et al.'s longest MEGAHIT contig happened to be the contig for SARS2 by chance, because there were no bacterial contigs longer than 30,000 bases even though one bacterial contig listed in Supplementary Table 1 was longer than 10,000 bases. When I ran MEGAHIT for Wu et al.'s raw reads, my second-longest contig was a 16,036-base contig for Leptotrichia hongkongensis and my third-longest contig was a 13,656-base contig for Veillonella parvula. However in other cases MEGAHIT will of course produce longer contigs for bacteria, and for example when I used MEGAHIT to assemble metagenomic reads of the bat sarbecovirus RmYN02, my longest contig was a contig for E. coli which was about 50,000 bases long, and the contig for RmYN02 was only the 4th-longest contig.
When I ran MEGAHIT Wu et al.'s raw reads, I got a total of about 30,000 contigs, but when I aligned my contigs against a set of about 15,000 virus reference sequences, I got only 8 contigs which matched viruses. I got only one contig which matched SARS2. And I got 4 contigs which matched the human endogenous retrovirus K113, but the contigs probably just came from the human genome since the sequences of HERVs are incorporated into the human genome. And I got one short contig which matched a Streptococcus phage and two short contigs which matched a parvovirus, both of which also had a handful of reads in the STAT results so they probably represent genuine matches to viruses. So among approximately 30,000 contigs, there may have been only 4 contigs which came from viruses. The reason why my number of contigs was an order of magnitude lower than the number reported by Wu et al. is probably because almost all human reads were masked with N bases in the reads uploaded to SRA, so my contigs included only a small number of contigs for fragments of the human genome.
About half of Wu et al.'s reads consist of only N bases, but it's probably human reads were masked because of NCBI's privacy policy. The NCBI has even published a command-line utility called Human Scrubber, which is used to replace human reads with N bases in raw reads that are submitted to the Sequence Read Archive. Wu et al. wrote that about 24 million reads out of about 57 million reads remained after they filtered out human reads, which roughly matches the reads at the SRA where there's about 27 million reads that consist of only N bases out of a total of about 57 million reads.
The first version of Wuhan-Hu-1 submitted to GenBank was 30,474 bases long but the current version is only 29,903 bases long. That's because the first version accidentally included a 618-base segment of human DNA at the 3' end, where the first 20 bases of the segment are part of both the actual genome of SARS2 and the human genome. The error was soon fixed in the second version of Wuhan-Hu-1 which was published at GenBank two days after the first version, and the error was even mentioned by Eddie Holmes on Twitter. I have found similar assembly errors in other sarbecovirus sequences, like RfGB02, Sin3408, Rs7907, Rs7931, GX-P1E, and Ra7909. So the reason why the original 30,474-base contig cannot be reproduced from the reads at the SRA is because almost all human reads were masked with N bases at SRA.
When the anonymous mathematician from Hamburg used BBMap to align Wu
et al.'s raw reads against the reference genomes of SARS2 and other
viruses, he got a large number of reads to align against the reference
genomes of HIV-1, measles, and hepatitis delta, but that's because he
ran BBMap with the parameter minratio=0.1 which used
extremely loose alignment criteria. Among the reads which aligned
against HIV-1, the average error rate was about 40% and the most common
read length 37 bases, but almost all of the reads would've remained
unaligned if he would've ran BBMap with the default parameters which
tolerate a much lower number of errors. The error rate was not mentioned
in his paper but I was able to calculate it by running his code myself.
However for the reads which aligned against SARS2, the most common
length was 151 bases and the average error rate of the 151-base reads
was only about 2%.
In the paper by the mathematician from Hamburg, there's plots for each virus reference sequence which show how many reads of each length aligned against the reference. The plot for SARS2 has a bimodal distribution with one peak around length 151 and another peak around length 35-40, but the plots for other viruses only have a single peak around length 35-40, which consists of reads which did not actually match the reference genome and which would have remained unaligned if he ran BBMap with the default parameters. But in the plot for SARS2, the y-axis is cut off in a deceptive way so you cannot see that the peak for reads around length 151 is much higher than the peaks around length 35-40 in the plots for other viruses.
Wu et al. wrote that the longest contig they got with Trinity was
only 11,760 bases long, but that's because Trinity split the genome of
SARS2 into multiple shorter contigs, which probably either overlapped or
had only small gaps in between. In order to merge the contigs, Wu et
al. could've aligned all contigs against a related virus like ZC45 or
SARS1, and they could've selected the most common base at each spot of
the alignment. And if any gaps remained, Wu et al. could've filled in
the gaps by using segments on either end of the gap as PCR primers and
by sequencing the PCR amplicons. By default Trinity and MEGAHIT both
require a region of a genome to be covered by at least two contigs
before the contigs get merged, so Wu et al. may have also gotten Trinity
to merge the shorter contigs into a single contig if they would've added
the option --min_glue 1 which reduces the minimum coverage
requirement from 2 to 1.
When I ran Trinity on Wu et al.'s raw reads without trimming, my longest contig was 29,879 bases long, and it was otherwise identical to the third version of Wuhan-Hu-1 except it included a 5-base insertion at the 5' end, it had one nucleotide change in the middle, and it was missing the last 29 bases of the poly(A) tail. When I ran Trinity again after trimming the reads, my longest contig was 31,241 bases long because there was a 1,379-base insertion in the middle which consisted of two segments that were identical to two different parts of Wuhan-Hu-1. The reason why Wu et al. failed to get a single complete contig with Trinity may have been because they used a different method to trim the reads, or because they used a different version of Trinity, or because Trinity was confused by the human reads which had been masked in my Trinity run.
In the current 29,903-base version of Wuhan-Hu-1, the poly(A) tail is 33 bases long, but the full poly(A) tail cannot be assembled from the metagenomic raw reads because the full poly(A) tail is not included in the raw reads. Wu et al. wrote that they sequenced the ends of the genome with RACE (rapid amplification of cDNA ends), which is a method similar to PCR which only requires specifying a single primer instead of two primers, so it can be used to amplify the end of a genome by specifying a single segment near the end as the primer. In October 2020 when the sequence of RaTG13 was updated at GenBank so that a new 15-base segment was added to the 5' end, some people from DRASTIC were suspecting fraud because the 15-base segment was not included in the metagenomic raw reads for RaTG13, but the segment was however included in the RACE amplicon sequences for RaTG13 which were only published later in 2021. But I believe the RACE amplicon sequences of Wuhan-Hu-1 have never been published.
When Wu et al. wrote that they determined the viral genome organization of WHCV by aligning it against SARS-CoV Tor2 and bat SL-CoVZC45, they meant that after they had already assembled the complete genome of SARS2, they aligned it against SARS1 and ZC45 in order to make annotating the proteins easier. So if for example they got one open reading frame that started around position 21,500 and ended around position 25,000, they knew that it was the spike protein if it covered roughly the same region as the spike protein of SARS1. But they did not mean that they aligned the raw reads of SARS2 against a SARS1 genome in order to do reference-based assembly.
USMortality did an experiment where he downloaded the reference genomes of various viruses, he generated simulated short reads for the genomes, and he tried to assemble the reads back together with MEGAHIT, but he failed to get complete contigs for a couple of viruses, like HKU1, HIV-1, HIV-2, and porcine adenovirus. But that's because the reference genome of HKU1 contains a region where the same 30-base segment is repeated 14 times in a row. And in HIV-1 and HIV-2 there's a long terminal repeat where a long segment at the 5' end of the genome is repeated at the 3' end of the genome. And porcine adenovirus contains a tandem repeat where the same 724-base segment is repeated twice in a row. Also when USMortality did another experiment where he mixed together simulated reads from multiple different viruses and he ran MEGAHIT on the mixed reads, he failed to get complete contigs for SARS2 and SARS1, but that's because the contigs were split at a spot where there's a 74-base segment that is identical in the reference genomes of SARS2 and SARS1, and when I repeated his experiment, I was able to get complete contigs for SARS2 and SARS1 when I simply increased the maximum k-value of MEGAHIT from 141 to 161.
One reason why USMortality says that the genome has not been validated is that in all of Wu et al.'s reads which align against the very start of the 5' end of Wuhan-Hu-1, there are 3-5 extra bases at the start of the read that usually end with one or more G bases. However the G bases are part of the template-switching oligo which anneals to complementary C bases that are added to the 5' end of the RNA strand by reverse transcriptase. USMortality also got two extra G bases at the start of one of his MEGAHIT contigs, but I was able to get rid of them by trimming 5 bases from the ends of reads.
The standard reference genome of SARS2 is known as Wuhan-Hu-1, and it was described in a paper by Wu et al. titled "A new coronavirus associated with human respiratory disease in China". [https://www.nature.com/articles/s41586-020-2008-3] There's currently three versions of Wuhan-Hu-1 at GenBank: MN908947.1 is 30,474 bases long and it was published on January 12th 2020, MN908947.2 is 29,875 bases long and it was published on January 14th 2020, and MN908947.3 is 29,903 bases long and it was published on January 17th 2020. [https://www.ncbi.nlm.nih.gov/nuccore/MN908947.3?report=girevhist] I believe the dates are in UTC and not in a local timezone.
Apart from a single nucleotide change in the N gene, the only differences between the three versions are at the beginning of the genome in the 5' UTR and at the end of the genome in the 3' UTR:
$ brew install mafft
[...]
$ curl -s 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=MN908947.'{1,2,3}>wuhu.fa
$ mafft --globalpair --clustalout --thread 4 wuhu.fa
[...]
CLUSTAL format alignment by MAFFT G-INS-1 (v7.490)
MN908947.1 cggtgacgcatacaaaacattcccaccataccttcccaggtaacaaaccaaccaactttc
MN908947.2 cggtgacgcatacaaaacattcccaccataccttcccaggtaacaaaccaaccaactttc
MN908947.3 -----------attaaaggtt-----tataccttcccaggtaacaaaccaaccaactttc
*. *** .** .*********************************
[... 29,160 bases omitted]
MN908947.1 tgacctacacagctgccatcaaattggatgacaaagatccaaatttcaaagatcaagtca
MN908947.2 tgacctacacagctgccatcaaattggatgacaaagatccaaatttcaaagatcaagtca
MN908947.3 tgacctacacaggtgccatcaaattggatgacaaagatccaaatttcaaagatcaagtca
************ ***********************************************
[... 480 bases omitted (part of N gene, whole ORF10, part of 3' UTR)]
MN908947.1 aatgtgtaaaattaattttagtagtgctatccccatgtgattttaatagcttcttcctgg
MN908947.2 aatgtgtaaaattaattttagtagtgctatccccatgtgattttaatagcttctt-----
MN908947.3 aatgtgtaaaattaattttagtagtgctatccccatgtgattttaatagcttcttaggag
*******************************************************
MN908947.1 gatcttgatttcaacagcctcttcctcatactattctcaacactactgtcagtgaacttc
MN908947.2 ------------------------------------------------------------
MN908947.3 aatgacaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaa---------------------
MN908947.1 taaaaatggattctgtgtttgaccgtaggaaacatttcagtattccttatctttagaaac
MN908947.2 ------------------------------------------------------------
MN908947.3 ------------------------------------------------------------
MN908947.1 ctctgcaactcaagtgtctctgccaaagagtcttcaaggtaatgtcaaataccataacat
MN908947.2 ------------------------------------------------------------
MN908947.3 ------------------------------------------------------------
MN908947.1 attttttaagtttttgtatacttcctaggaatatatgtcatagtatgtaaagaagatctg
MN908947.2 ------------------------------------------------------------
MN908947.3 ------------------------------------------------------------
MN908947.1 attcatgtgatctgtactgttaagaaacacttcaaaggaaaacaacctctcatagatctt
MN908947.2 ------------------------------------------------------------
MN908947.3 ------------------------------------------------------------
MN908947.1 caaatcctgttgttcttgagttggaatgtacaatattcaggtagagatgctggtacaaaa
MN908947.2 ------------------------------------------------------------
MN908947.3 ------------------------------------------------------------
MN908947.1 aaagactgaatcatcaggctattagaaagataaactaggcctctaacatagtaactttaa
MN908947.2 ------------------------------------------------------------
MN908947.3 ------------------------------------------------------------
MN908947.1 cagtttgtacttatacatattttcacattgaaatatagttttattcatgactttttttgt
MN908947.2 ------------------------------------------------------------
MN908947.3 ------------------------------------------------------------
MN908947.1 tttagcttctctgtcttccattatttcaagctgctaaaaattaaaaatatcctatagcaa
MN908947.2 ------------------------------------------------------------
MN908947.3 ------------------------------------------------------------
MN908947.1 agggctatggcatctttttgtaaaaataaggaaagcaaggttttttgataatc
MN908947.2 -----------------------------------------------------
MN908947.3 -----------------------------------------------------
The first and second versions started with the 27-base segment
CGGTGACGCATACAAAACATTCCCACC which was changed to
ATTAAAGGTTT in the third version. If you remove the first 3
bases from CGGTGACGCATACAAAACATTCCCACC, then it's identical
to a 24-base segment that's included near the end of all three versions
Wuhan-Hu-1:
$ seqkit grep -nrp \\.1 wuhu.fa|seqkit subseq -r4:27|seqkit locate -f- wuhu.fa|cut -f1,4-|column -t seqID strand start end matched MN908947.1 + 4 27 TGACGCATACAAAACATTCCCACC MN908947.1 + 29360 29383 TGACGCATACAAAACATTCCCACC MN908947.2 + 4 27 TGACGCATACAAAACATTCCCACC MN908947.2 + 29360 29383 TGACGCATACAAAACATTCCCACC MN908947.3 + 29344 29367 TGACGCATACAAAACATTCCCACC
I have found similar assembly errors in several other sarbecovirus sequences, where there's a short segment at either end of the sequence which consists of another part of the genome or its reverse complement. For exaple in a sequence for the SARS1 isolate SoD, there's a 24-base segment at the 3' end which is the reverse complement of another 24-base segment near the 3' end:
$ curl 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=AY461660.1'>sod.fa $ seqkit locate -ip cacattagggctcttccatatagg sod.fa|cut -f4-|column -t strand start end matched + 29692 29715 cacattagggctcttccatatagg - 29629 29652 cacattagggctcttccatatagg $ seqkit subseq -r 29629:29652 sod.fa # the match on the minus strand is a reverse complement of the match on the plus strand >AY461660.1 SARS coronavirus SoD, complete genome CCTATATGGAAGAGCCCTAATGTG
Assembly errors like this are easy to spot if you do a multiple sequence alignment of related virus sequences, and you then look for inserts at the beginning or end of the alignment which are only included in one sequence but missing from all other sequences. And you can then do a BLAST search for the insert to find its origin.
The 3' end of the first version of Wuhan-Hu-1 accidentally included a 618-base segment of human DNA. [https://x.com/ChrisDeZPhD/status/1290218272705531905] The first 20 of the 618 bases are part of the actual genome of SARS2 which may have confused MEGAHIT. You can do a BLAST search for the 618-base segment by going here: https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn. Then paste the sequence below to the big text field at the top and press the "BLAST" button:
$ curl -s 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=MN908947.1'|seqkit subseq -r -618:-1 >MN908947.1 Wuhan seafood market pneumonia virus isolate Wuhan-Hu-1, complete genome tgtgattttaatagcttcttcctgggatcttgatttcaacagcctcttcctcatactatt ctcaacactactgtcagtgaacttctaaaaatggattctgtgtttgaccgtaggaaacat ttcagtattccttatctttagaaacctctgcaactcaagtgtctctgccaaagagtcttc aaggtaatgtcaaataccataacatattttttaagtttttgtatacttcctaggaatata tgtcatagtatgtaaagaagatctgattcatgtgatctgtactgttaagaaacacttcaa aggaaaacaacctctcatagatcttcaaatcctgttgttcttgagttggaatgtacaata ttcaggtagagatgctggtacaaaaaaagactgaatcatcaggctattagaaagataaac taggcctctaacatagtaactttaacagtttgtacttatacatattttcacattgaaata tagttttattcatgactttttttgttttagcttctctgtcttccattatttcaagctgct aaaaattaaaaatatcctatagcaaagggctatggcatctttttgtaaaaataaggaaag caaggttttttgataatc
The best match is 616 bases long because it's missing the last 2 bases of the query, but 614 out of 616 bases are identical with a result titled "Human DNA sequence from clone RP11-173E24 on chromosome 1q31.1-31.3, complete sequence".
I have also found other sarbecovirus sequences where a piece of host DNA or rRNA was accidentally included at either end of the genome. For example in a sequence for the SARS-like bat virus Rs7907, I noticed that the 5' end had an insertion which was missing from other SARS-like viruses:
$ curl https://sars2.net/f/sarbe.fa.xz|xz -dc>sarbe.fa
$ seqkit fx2tab sarbe.fa|grep -Ev 'Rs7931|Ra7909'|grep -C10 Rs7907|awk -F\\t '{print$1 FS substr($2,1,550)}'|seqkit tab2fx|mafft --globalpair --quiet -|seqkit fx2tab|awk -F\\t '{print $2 FS$1}'|cut -d, -f1|column -ts$'\t'
a----------------------------------------------------------------taaaaggattcatccttccc---- MT726044.1 Bat coronavirus isolate PREDICT/PDF-2370/OTBA35RSV
a----------------------------------------------------------------taaaaggattcatccttccc---- MT726043.1 Sarbecovirus sp. isolate PREDICT/PDF-2386/OTBA40RSV
-----------------------------------------------------------------taaaaggattaatccttccc---- KY352407.1 Severe acute respiratory syndrome-related coronavirus strain BtKY72
----------------------------------------------------------------------------------------- NC_014470.1 Bat coronavirus BM48-31/BGR/2008
----------------------------------------------------------------------------------------- GU190215.1 Bat coronavirus BM48-31/BGR/2008
----------------------------------------------------------------------------------------- MZ190137.1 Bat SARS-like coronavirus Khosta-1 strain BtCoV/Khosta-1/Rh/Russia/2020
----------------------------------------------------------------------------------------- OL674081.1 Severe acute respiratory syndrome-related coronavirus isolate Rs7952
----------------------------------------------------------------------------------------- OL674074.1 Severe acute respiratory syndrome-related coronavirus isolate Rs7896
-----------------------------------------------------------------------------accttcccaggt OL674075.1 Severe acute respiratory syndrome-related coronavirus isolate Rs7905
-------------------------------------------------------------------------------cttcccaggt OL674079.1 Severe acute respiratory syndrome-related coronavirus isolate Rs7924
ggggattgcaattattccccatgaacgaggaattcccagtaagtgcgggtcataagcttgcgttgattaagtccctgccctttgtacac OL674076.1 Severe acute respiratory syndrome-related coronavirus isolate Rs7907
------------------------------------------------------------------------------ccttcccaggt OL674078.1 Severe acute respiratory syndrome-related coronavirus isolate Rs7921
a----------------------------------------------------------------ttaaaggtttttaccttcccaggt MZ081380.1 Betacoronavirus sp. RsYN04 strain bat/Yunnan/RsYN04/2020
a----------------------------------------------------------------ttaaaggtttttaccttcccaggt MZ081378.1 Betacoronavirus sp. RmYN08 strain bat/Yunnan/RmYN08/2020
a----------------------------------------------------------------ttaaaggtttttaccttcccaggt MZ081376.1 Betacoronavirus sp. RmYN05 strain bat/Yunnan/RmYN05/2020
When I did a BLAST search for the first 150 bases of the Rs7907 sequence, the first 96 bases were 100% identical with several sequences of mammalian DNA or rRNA, like for example there were results titled "PREDICTED: Pan troglodytes 18S ribosomal RNA (LOC129143085), rRNA" and "Delphinus delphis genome assembly, chromosome: 21".
In the paper where Rs7907 was published, the authors didn't describe removing host reads before they did de-novo assembly: "For SARSr-CoV-positive RNA extraction, next-generation sequencing (NGS) was performed using BGI MGISEQ 2000. NGS reads were first processed using Cutadapt (v.1.18) to eliminate possible contamination. Thereafter, the clean reads were assembled into genomes using Geneious (v11.0.3) and MEGAHIT (v1.2.9). PCR and Sanger sequencing were used to fill the genome gaps. To amplify the terminal ends, a SMARTer RACE 5'/3'kit (Takara) was used." [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8344244/] (However I don't know if they did RACE for the 5' tail of Rs7907, because they would've noticed the assembly error in their contig if they would've incorporated the RACE sequences into their contig, or maybe they just published their MEGAHIT contig without combining it with the RACE sequences.)
The assembly error in the first version of Wuhan-Hu-1 was also mentioned by Eddie Holmes, who tweeted that "Zhang's sequence did have some human sequences at the ends which we quickly fixed". [https://x.com/edwardcholmes/status/1643722487333789696] And in an email to USMortality, Eddie Holmes also wrote: "I do know that the initial upload to Virological/GenBank had some human DNA at the 5' and 3' ends that was then corrected." [https://usmortality.substack.com/p/why-do-wu-et-al-2020-refuse-to-answer] However the insert at the 5' end did not actually come from the human genome but from another part of the genome of SARS2.
When the middle part of an RNA sequence is known but it's not certain if the ends of the sequence are correct, the ends can be sequenced accurately using method called RACE (rapid amplification of cDNA ends). RACE is also known as "one-sided PCR" or "anchored PCR", and it basically requires only specifying a single primer for each amplified segment instead of a forward and reverse primer like in regular PCR. [https://en.wikipedia.org/wiki/Rapid_amplification_of_cDNA_ends]
In the third version of Wuhan-Hu-1 at GenBank, an additional 44 bases were inserted to the 3' end so that the poly(A) tail became 33 bases long. I believe it's because the ends of the genome were sequenced with RACE, because the paper by Wu et al. said: "The genome sequence of this virus, as well as its termini, were determined and confirmed by reverse-transcription PCR (RT-PCR)10 and 5'/3' rapid amplification of cDNA ends (RACE), respectively." [https://www.nature.com/articles/s41586-020-2008-3]
Supplementary Table 6 shows the primers used to do RACE, where on each row the number in parentheses is the expected length of the amplicon which extends from the primer until the end of the genome: [https://static-content.springer.com/esm/art%3A10.1038%2Fs41586-020-2008-3/MediaObjects/41586_2020_2008_MOESM1_ESM.pdf]
| D. Primers used in 5'/3' RACE | |||
| 5-GSP | CCACATGAGGGACAAGGACACCAAGTG | 573-599 (599 bp) | |
| 5-GSPn | CATGACCATGAGGTGCAGTTCGAGC | 491-515 (515 bp) | |
| 3-GSP | TGTCGCGCATTGGCATGGAAGTCACACC | 29212-29239 (688 bp) | |
| 3-GSPn | CTCAAGCCTTACCGCAGAGACAGAAG | 29398-29423 (502 bp) | |
In 2020, Steven Quay posted a tweet where he wrote: "Dr. Shi lied? Below is NEW RaTG13 sequence https://ncbi.nlm.nih.gov/nuccore/MN996532 with the missing 1-15 nt at 5' end. So I blasted 1-180 of this new sequence against her RNA-Seq for RaTG13 to find the missing 15 nt. But they are not there. So 1-15 nt appears FABRICATED by Dr. Shi & WIV." [https://x.com/quay_dr/status/1318041151211884552] Actually the RACE sequences of RaTG13 were only added to SRA in December 2021 after Quay's tweet, but one of them matched the segment of 15 nucleotides which was added to the start of the second version of RaTG13: [https://www.ncbi.nlm.nih.gov/sra/?term=SRR16979454]
$ fastq-dump SRR16979454 # RACE sequences of RaTG13
$ curl 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=MN996532.'{1,2}|mafft ->ratg.fa # first and second version of RaTG13
[...]
$ aldi2(){ cat "$@">/tmp/aldi;Rscript --no-init-file -e 'm=t(as.matrix(Biostrings::readAAStringSet("/tmp/aldi")));diff=which(rowSums(m!=m[,1])>0);split(diff,cumsum(diff(c(-Inf,diff))!=1))|>sapply(\(x)c(if(length(x)==1)x else paste0(x[1],"-",tail(x,1)),apply(m[x,,drop=F],2,paste,collapse="")))|>t()|>write.table(quote=F,sep=";",row.names=F)';}
;MN996532.1 Bat coronavirus RaTG13, complete genome;MN996532.2 Bat coronavirus RaTG13, complete genome
1-15;---------------;attaaaggtttatac
2125;c;t
2132;c;a
6936;c;a
7127;g;a
18812-18813;gc;tg
29856-29870;aaaaaaaaaaaaaaa;---------------
$ seqkit locate -irp attaaaggtttatac SRR16979454.fastq|column -ts$'\t' # search RACE sequences for the 15-base segment that was added to the start of the second version of RaTG13
seqID patternName pattern strand start end matched
SRR16979454.4 attaaaggtttatac attaaaggtttatac - 461 475 ATTAAAGGTTTATAC
As far as I know, the RACE sequences used by Wu et al. were never published, but I guess they match a longer part of the poly(A) tail than the metagenomic reads which only included a few bases of the poly(A) tail.
All five BANAL viruses are also missing parts of the 5' UTR like the first version of RaTG13:
$ curl -sL sars2.net/f/sarbe.fa.xz|xz -dc>sarbe.fa
$ wget sars2.net/f/sarbe.pid
$ curl -s 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=MN996532.1'|sed '1s/.*/>RaTG13_version_1_no_RACE/'>ratg1.fa
$ awk -F\\t 'NR==1{for(i=2;i<=NF;i++)if($i~/Wuhan-Hu-1/)break;next}$i>93{print$1}' sarbe.pid|cut -d\ -f1|seqkit grep -f- sarbe.fa|cat - ratg1.fa|seqkit subseq -r1:1000|mafft --quiet --thread 4 -|seqkit subseq -r 1:60|seqkit fx2tab|sed $'s/.* \\(.*\\),[^\t]*/\\1/'|column -t
Wuhan-Hu-1 attaaaggtttataccttcccaggtaacaaaccaaccaactttcgatctcttgtagatct
BANAL-20-236/Laos/2020 -ttaaaggtttataccttcccaggtaacaaaccaaccaactctcgatctcttgtagatct
BANAL-20-103/Laos/2020 -----------------------------------------ctcgaattcttgtagatct
BANAL-20-52/Laos/2020 ------------------------taacaaaccaaccaactttcgatctcttgtagatct
RaTG13 attaaaggtttatacctttccaggtaacaaaccaacgaactctcgatctcttgtagatct
bat/Yunnan/RpYN06/2020 attaaaggtttataccttcccaggtaacaaaccaaccaaccctcgatctcttgtagatct
BANAL-20-247/Laos/2020 --------------------------------------actctcgatctcttgtagatct
BANAL-20-116/Laos/2020 ------------------------------------------------------------
RaTG13_version_1_no_RACE ---------------ctttccaggtaacaaaccaacgaactctcgatctcttgtagatct
When I did local alignment of the metagenomic reads for RaTG13 against the second version of RaTG13 at GenBank, the minimum starting position among the aligned reads was only 15:
$ curl -s 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=MN996532'>ratg13.fa
$ parallel-fastq-dump --gzip --threads 10 --split-files --sra-id SRR11085797
[...]
$ minimap2 -a --sam-hit-only ratg13.fa SRR11085797_[12].fastq.gz|awk '$10=="*"{$10=x}$11=="*"{$11=y}{x=$10;y=$11}1' {,O}FS=\\t|samtools sort -@3 ->ratg13.bam
[...]
$ samtools view ratg13.bam|head -n4|cut -f1,2,4,6,10,12|tr \\t \|
SRR11085797.8084709|0|15|151M|CCTTTCCAGGTAACAAACCAACGAACTCTCGATCTCTTGTAGATCTGTTCTCTAAACGAACTTTAAAATCTGTGTGACTGTCACTCGGCTGCATGCTTAGTGCACTCACGCAGTATAATTAATAACTAATTACTGTCGTTGACAGGACACG|NM:i:0
SRR11085797.8811160|0|16|150M|CTTTCCAGGTAACAAACCAACCAACTCTCGATCTCTTGTAGATCTGTTCTCTAAACGAACTTTAAAATCTGTGTGACTGTCACTCGGCTGCATGCTTAGTGCACTCACGCAGTATAATTAATAACTAATTACTGTCGTTTACAGGACACG|NM:i:2
SRR11085797.8084709|16|18|150M|TTCCAGGTNACAAACCAACGAACTCTCGATCTCTTGTAGATCTGTTCTCTAAACGAACTTTAAAATCTGTGTGACTGTCACTCGGCTGCATGCTTAGTGCACTCACGCAGTATAATTAATAACTAATTACTGTCGTTGACAGGACACGAG|NM:i:1
SRR11085797.3139935|0|25|143M|TAACAAACCAACCAACTCTCGATCTCTTGTAGATCTGTTCTCTAAACGAACTTTAAAATCTGTGTGACTGTCACTCGGCTGCATGCTTAGTGCACTCACGCAGTATAATTAATAACTAATTACTGTCGTTGACAGGACACGAG|NM:i:1
The third version of Wuhan-Hu-1 has a 33-base poly(A) tail, but I think they sequenced the tail with RACE because there's only a few bases of the tail included in the metagenomic reads published at the SRA. The output below shows reads that aligned against the end of the third version of Wuhan-Hu-1. To save space, I only included one read per each starting position even though many positions have over ten aligned reads. The first column shows the starting position and the second column shows the number of mismatches relative to Wuhan-Hu-1:
$ brew install bowtie2 fastp samtools
[...]
$ wget ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR109/081/SRR10971381/SRR10971381_{1,2}.fastq.gz
[...]
$ curl -s 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=MN908947.3'>sars2.fa
$ bowtie2-build sars2.fa{,}
[...]
$ fastp -i SRR10971381_1.fastq -I SRR10971381_2.fastq -o SRR10971381_1.trim.fq.gz -O SRR10971381_2.trim.fq.gz -l 70
[...]
$ bowtie2 -p3 -x sars2.fa -1 SRR10971381_1.trim.fq.gz -2 SRR10971381_2.trim.fq.gz --no-unal|samtools sort -@2 ->sorted58.bam
[...]
$ samtools view sorted58.bam|awk -F\\t '!a[$4]++{x=$4;y=$10;sub(/.*NM:i:/,"");print x,$1,y}'|tail -n30
29763 1 AATGAACAATGCTAGGGAGAGCTGCCTATATGGAAGAGCCCTAATGTGTAAAATTAATTTTAGTAGTGCTATCCCCATGTGATTTTAATAGCTTCTTAGGAG
29767 2 TTCAATGCTAGGGAGAGCTGCCTATATGGAAGAGCCCTAATGTGTAAAATTAATTTTAGTAGTGCTATCCCCATGTGATTTTAATAGCTTCTTAGGAGAA
29769 8 GGCTGCTAGGGAGAGCTGCCTATATGGAAGAGCCCTAATGTGTAAAATTAATTTTAGTAGTGCTATCCCCATGTGATTTTAATAGCGTCTCTTTAGA
29771 4 GGTCTAGGGAGAGCTGCCTATATGGAAGAGCCCTAATGTGTAAAATTAATTTTAGTAGTGCTATCCCCAA
29772 2 TTTTAGGGAGAGCTGCCTATATGGAAGAGCCCTAATGTGTAAAATTAATTTTAGTAGTGCTATCCCCATGTGATTTTAATAGCTTCTTAGGAGAA
29773 1 GTTAGGGAGAGCTGCCTATATGGAAGAGCCCTAATGTGTAAAATTAATTTTAGTAGTGCTATCCCCATGTGATTTTAATAGCTTCTTAGGAGA
29775 2 TATGGAGAGCTGCCTATATGGAAGAGCCCTAATGTATAAAATTAATTTTAGTAGTGCTATCCCCATGTGATTTTAATAGCTTCTTAGGAGAATGACA
29776 6 TTGGAGAGCTGCCTATATGGCAGAGCCCTAATGTGTAAAATTAATTTTAGTAGTGCTATCCCCATGTGATTTTAATAGCTTCTTAGATGC
29777 2 ATGAGAGCTGCCTATATGGAAGAGCCCTAATGTGTAAAATTAATTTTAGTAGTGCTATCCCCATGTGATTTTAATAGCTTCTTAGGAG
29780 1 AGTGCTGCCTATATGGAAGAGCCCTAATGTGTAAAATTAATTTTAGTAGTGCTATCCCCATGTGATTTTAATAGCTTCTTAGG
29781 4 AAACTGCCTATATGGAAGAGCCCTAATGTGTAAAATTAATTTTAGTAGTGCTATCCGCATGTGATTTTAATAGCTTCTTAGGAGAATAACA
29791 1 TGTGGAAGAGCCCTAATGTGTAAAATTAATTTTAGTAGTGCTATCCCCATGTGATTTTAATAGCTTCTTAGGAGAA
29855 8 CTGTTAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
29857 8 AATAAAAAAAACAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
29858 8 AAAAAAAAAACTCAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
29859 6 AAAAAGAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
29860 7 AAATAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
29861 6 AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
29862 3 AAAAATCACAAAAAAAAAAAAAAAAAAAAAAAA
29863 4 AAAAAAATAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
29864 4 AAACAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
29865 1 AATGAAAAAAAAAAAAAAAAAAAAAAAAAAA
29866 3 AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
29867 4 AAAAAAAAAATAAAAAAAAAAAAAAAAAAAAAAAAAA
29868 2 AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
29869 1 AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
29870 1 AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
29871 0 AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
29872 1 AATAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
29873 1 AAAAAATAAAAAAAAAAAAAAAAAAAAAAAA
In the output above, there's a huge gap between positions 29,791 and 29,855 with no reads starting from the 63 positions in between. I think it's because the reads after the gap don't actually come from SARS2 but from some other organisms.
The last 50 bases of Wuhan-Hu-1 before the poly(A) tail are
ATTTTAGTAGTGCTATCCCCATGTGATTTTAATAGCTTCTTAGGAGAATGAC, which
is matched in the by the reads above the gap within a couple of
mismatches. But the first read below the gap starts with
CTGTT which has three nucleotide changes from
ATGAC at the corresponding position in Wuhan-Hu-1.
Out of the reads whose starting position is 29,700 or above, there's
only one read which has five A bases after the sequence
ATGAC:
$ samtools view sorted58.bam|awk '$4>=29700'|cut -f10|grep -Eo ATGACA+|sort|uniq -c|sort -n
1 ATGACAAAAA
2 ATGACAAA
2 ATGACAAAA
8 ATGACA
I'm not sure which of the dates above are UTC and which are local time.
In Supplementary Table 8 of Wu et al. 2020, there's a list of the PCR
primers they used to do PCR-based sequencing in order to verify the
results of the metagenomic de-novo assembly they did with MEGAHIT.
[https://static-content.springer.com/esm/art%3A10.1038%2Fs41586-020-2008-3/MediaObjects/41586_2020_2008_MOESM1_ESM.pdf]
The last reverse primer is AAAATCACATGGGGATAGCACTACT which
is shown to end at position 29,864, so I guess I guess by the time they
designed the primers, they had already noticed that they accidentally
included a piece of human DNA at the beginning of the first version of
Wuhan-Hu-1 at GenBank. The first forward primer is
CCAGGTAACAAACCAACCAACTT and it's shown to start at position
36, which matches the coordinates in the first and second version of
Wuhan-Hu-1 but not the third version, so I guess they designed the
primers based on the second version. However another part of
Supplementary Table 8 says that the "primers for
WHCV detection using qPCR" were "designed
based on the whole genome of WHCV (MN908947.3)", and the
coordinates of the primers match the third version of Wuhan-Hu-1 at
GenBank and not the second version. So I guess Wu et al. designed
different sets of primers based on different versions of the genome.
Below I ran MEGAHIT for Wu et al.'s lung metagenomic reads. When I
did trimming with fastp, my longest contig was 29,802 bases
long and it missed the last 102 bases of MN908947.3 and had an insertion
of one base at the beginning. When I did trimming with
trimmomatic, my longest contig was 29,875 bases long and
now it only missed the last 30 bases of MN908947.3 even though it still
had the single-base insertion at the beginning:
brew install megahit -s # `-s` compiles from source because the bottle was compiled against GCC 9 so it gave me the error `dyld: Library not loaded: /usr/local/opt/gcc/lib/gcc/9/libgomp.1.dylib`
wget ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR109/081/SRR10971381/SRR10971381_{1,2}.fastq.gz # download from ENA because it's faster than downloading from SRA and doesn't require installing sratoolkit
fastp -i SRR10971381_1.fastq.gz -I SRR10971381_2.fastq.gz -o SRR10971381_1.trim.fq.gz -O SRR10971381_2.trim.fq.gz -l 70
megahit -1 SRR10971381_1.trim.fq.gz -2 SRR10971381_2.trim.fq.gz -o megahutrim
brew install trimmomatic
trimmomatic PE SRR10971381_{1,2}.fastq.gz SRR10971381_{1,2}.paired.fq.gz SRR10971381_{1,2}.unpaired.fq.gz AVGQUAL:20 HEADCROP:12 LEADING:3 TRAILING:3 MINLEN:75 -threads 4 # adapter trimming was omitted here
megahit -1 SRR10971381_1.paired.fq.gz -2 SRR10971381_2.paired.fq.gz -o megahutrimmo
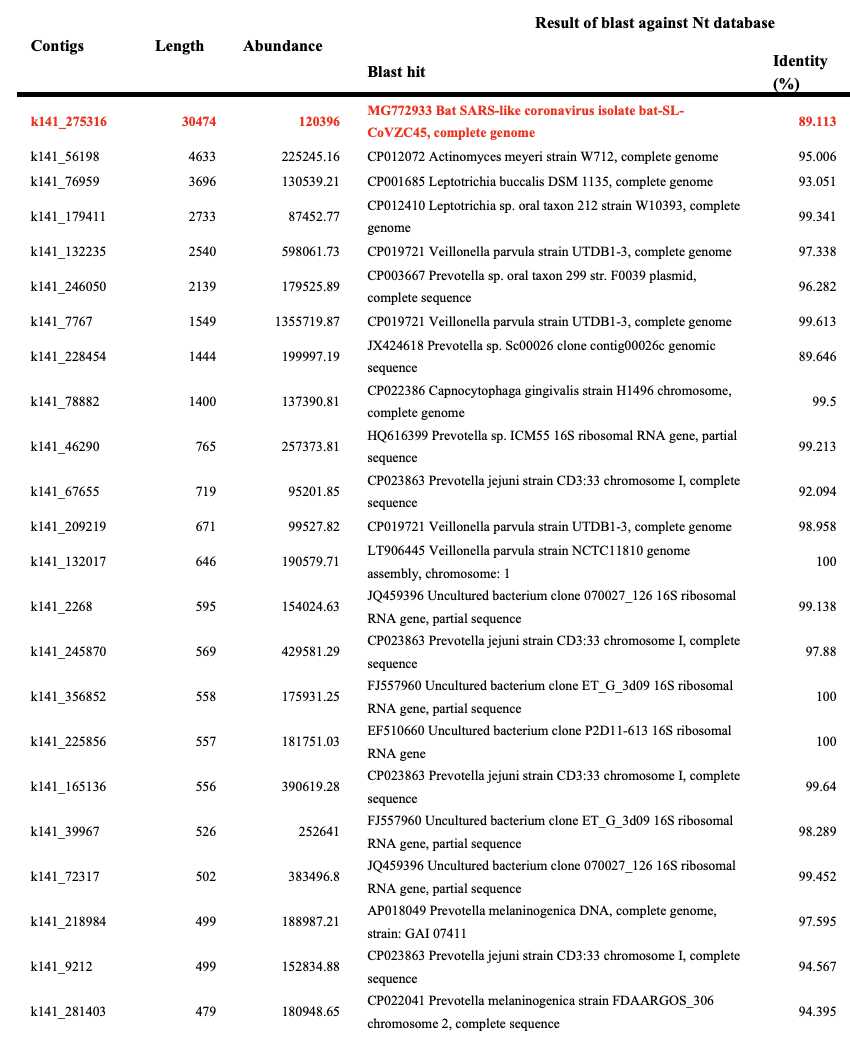
The paper by Wu et al. says: "In total, we generated 56,565,928 sequence reads that were de novo-assembled and screened for potential aetiological agents. Of the 384,096 contigs assembled by Megahit [9], the longest (30,474 nucleotides (nt)) had a high abundance and was closely related to a bat SARS-like coronavirus (CoV) isolate - bat SL-CoVZC45 (GenBank accession number MG772933) - that had previously been sampled in China, with a nucleotide identity of 89.1% (Supplementary Tables 1, 2).". And 56,565,928 is identical to the number of reads in the SRA run deposited by Wu et al.. [https://www.ncbi.nlm.nih.gov/sra/?term=SRR10971381] And 30,474 is identical to the length of the first version of Wuhan-Hu-1 at GenBank.
However when I tried aligning the reads in Wu et al.'s metagenomic dataset against the first version of Wuhan-Hu-1, I got zero coverage for the last 579 bases, and the following were the reads with the highest ending position:
$ curl -s 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=MN908947.1'>wuhu1.fa
$ bowtie2-build wuhu1.fa{,}
[...]
$ bowtie2 -p3 -x wuhu1.fa -1 SRR10971381_1.fastq.gz -2 SRR10971381_2.fastq.gz --no-unal|samtools sort -@2 ->sorted59.bam
[...]
$ samtools view sorted59.bam|ruby -alne'puts [$F[3],$F[3].to_i+$F[5].scan(/\d+(?=[MD])/).map(&:to_i).sum,$F[5],$F.grep(/NM:i/)[0][/\d+/],$F[9]]*" "'|sort -nk2|tail|column -t
29791 29887 96M 9 CGGGGAGAGCTGCCTATATGGAAGAGCCCTAATGTGTAAAATTAATTTTAGTAGTGCTATCCCCATGTGATTTTAATAGCTTCTTAGGAGAATGTC
29797 29888 91M 12 AAACTGCCTATATGGAAGAGCCCTAATGTGTAAAATTAATTTTAGTAGTGCTATCCGCATGTGATTTTAATAGCTTCTTAGGAGAATAACA
29766 29889 118M2I5M 13 GGCTCGAGTGTACAGTGAACAATGCTAGGGAGAGCTGCCTATATGGAAGAGCCCTAATGTGTAAAATTAATTTTAGTAGTGCTATCCCCATCTGATTTTAATAGCTTCTTAGGAGAATGACAAAA
29736 29890 139M3D12M 8 ACCACATTTTCACCGAGGCCACGCGGAGTACGATCGAGTGTACAGTGAACAATGCTAGGGAGAGCTGCCTATATGGAAGAGCCCTAATGTGTAAAATTAATTTTAGTAGTGCTATCCCCATGTGATTTTAATAGCTTCTTAGGAGAATGAC
29736 29890 139M3D12M 8 ACCACATTTTCACCGAGGCCACGCGGAGTACGATCGAGTGTACAGTGAACAATGCTAGGGAGAGCTGCCTATATGGAAGAGCCCTAATGTGTAAAATTAATTTTAGTAGTGCTATCCCCATGTGATTTTAATAGCTTCTTAGGAGAATGAC
29736 29890 139M3D12M 8 ACCACATTTTCACCGAGGCCACGCGGAGTACGATCGAGTGTACAGTGAACAATGCTAGGGAGAGCTGCCTATATGGAAGAGCCCTAATGTGTAAAATTAATTTTAGTAGTGCTATCCCCATGTGATTTTAATAGCTTCTTAGGAGAATGAC
29752 29890 132M2I6M 10 GGCCACGCGGAGTACGATCGAGTGTACAGTGAACAATGCTAGGGAGAGCTGCCTATATGGAAGAGCCCTAATGTGTAAAATTAATTTTAGTAGTGCTATCCCCATGTGATTTTAATAGCTTCTTAGGAGAATGACAAAAT
29791 29890 99M 11 TAAGGAGAGCTGCCTATATGGAAGAGCCCTAATGTGTAAAATTAATTTTAGTAGTGCTATCCCCATGTGATTTTAATAGCTTCTTAGGAGAATGACAAA
29766 29891 125M 13 TTATCGAGTGTACAGTGAACAATGCTAGGGAGAGCTGCCTATATGGAAGAGCCCTAATGTGTAAAATTAATTTTAGTAGTGCTATCCCCATGTGATTTTAATAGCTTCTTAGGAGAATGAGAAAA
29752 29895 123M3D17M 13 TGACACGCGGAGTACGATCGAGTGTACAGTGAACAATGCTAGGGAGAGCTGCCTATATGGAAGAGCCCTAATGTGTAAAATTAATTTTAGTAGTGCTATCCCCATGTGATTTTAATAGCTTCTTAGGAGAATGACAAAAA
In Wu et al.'s metagenomic dataset, about half of the reads have been
masked entirely so that they consist of only N bases:
$ seqkit grep -rsp '^N+$' SRR10971381_1.fastq.gz|seqkit head -n1 @SRR10971381.2 2 length=151 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN + !!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!! $ seqkit grep -rsp '^N+$' SRR10971381_1.fastq|seqkit stat # out of a total of about 28 million reads, about 14 million reads consist of only N letters file format type num_seqs sum_len min_len avg_len max_len - FASTQ DNA 13,934,971 2,027,338,447 35 145.5 151
So it could be that by the time Wu et al. submitted the reads to SRA, they had noticed that their initial 30,474-base contig included a segment of human DNA at the end, so maybe they replaced the reads which matched human DNA with reads that consisted of only N letters. That would explain why the number of reads at SRA is the same as the number of reads which produced their 30,474-base contig but why the contig cannot be reproduced. But it could be that at the point when they noticed the assembly error in MN908947.1, they were no longer able to update the main text of their paper in Nature, which might explain why the assembly error in MN908947.1 was accounted for in Supplementary Table 8 but not in the main text of the paper.
Wu et al. wrote that about 24 million reads out of about 57 million reads remained after human reads were filtered out:
Sequencing reads were first adaptor and quality trimmed using the Trimmomatic program[32]. The remaining 56,565,928 reads were assembled de novo using both Megahit (v.1.1.3)[9] and Trinity (v.2.5.1)[33] with default parameter settings. Megahit generated a total of 384,096 assembled contigs (size range of 200-30,474 nt), whereas Trinity generated 1,329,960 contigs with a size range of 201-11,760 nt. All of these assembled contigs were compared (using BLASTn and Diamond BLASTx) against the entire non-redundant (nr) nucleotide and protein databases, with e values set to 1 × 10−10 and 1 × 10−5, respectively. To identify possible aetiological agents present in the sequencing data, the abundance of the assembled contigs was first evaluated as the expected counts using the RSEM program[34] implemented in Trinity. Non-human reads (23,712,657 reads), generated by filtering host reads using the human genome (human release 32, GRCh38.p13, downloaded from Gencode) by Bowtie2[35], were used for the RSEM abundance assessment.
However in the STAT analysis for the reads uploaded to SRA, only
about 0.1% of the reads match Homo sapiens, about 0.8% of the
reads match simians, and about 4.6% of the reads match eukaryotes.
There's also an unusually low percentage of only 39% identified reads,
because the half of reads which consist of only N bases are
unidentified:
$ curl -s 'https://trace.ncbi.nlm.nih.gov/Traces/sra-db-be/run_taxonomy?cluster_name=public&acc=SRR10971381'>SRR10971381.stat $ jq -r '.[].tax_totals.total as$tot|.[].tax_table|sort_by(-.total_count)[]|(100*.total_count/$tot|tostring)+";"+.org' SRR10971381.stat|egrep 'Eukaryota|Simiiformes|Homo sapiens' 4.591841930004224;Eukaryota 0.8129416704698984;Simiiformes 0.1192095708215023;Homo sapiens $ jq -r '.[].tax_totals|100*.identified/.total' SRR10971381.stat 39.12950566284354
In order to compare the results to other human BALF samples from
patients with pneumonia, I searched the SRA for the phrase
bronchoalveolar pneumonia metagenomic human. In one run
which was part of a study titled "Bronchoalveolar
Lavage Fluid Metagenomic Next-Generation Sequencing in non-severe and
severe Pneumonia", about 86% of the reads were identified and 76%
matched simians:
[https://www.ncbi.nlm.nih.gov/sra/?term=SRR22183690]
$ curl -s 'https://trace.ncbi.nlm.nih.gov/Traces/sra-db-be/run_taxonomy?cluster_name=public&acc=SRR22183690'>SRR22183690.stat $ jq -r '.[].tax_totals.total as$tot|.[].tax_table|sort_by(-.total_count)[]|(100*.total_count/$tot|tostring)+";"+.org' SRR22183690.stat|egrep 'Eukaryota|Simiiformes|Homo sapiens' 81.79456158724525;Eukaryota 75.61440435123265;Simiiformes 12.85625010794863;Homo sapiens $ jq -r '.[].tax_totals|100*.identified/.total' SRR22183690.stat 86.16966070371517
In an early metagenomic sequencing run for SARS2 titled "Total RNA sequencing of BALF (human reads removed)", only about 0.6% of reads matched simians. The run was submitted by Wuhan University and published on January 18th 2020, and I didn't find any SARS2 run on SRA with an earlier publication date. [https://www.ncbi.nlm.nih.gov/bioproject/PRJNA601736] About 13% of the reads were masked so that they consisted of only N bases:
$ curl -s 'https://trace.ncbi.nlm.nih.gov/Traces/sra-db-be/run_taxonomy?cluster_name=public&acc=SRR10903402'>SRR10903402.stat $ jq -r '.[].tax_totals.total as$tot|.[].tax_table|sort_by(-.total_count)[]|(100*.total_count/$tot|tostring)+";"+.org' *02.stat|egrep 'Eukaryota|Simiiformes|Homo sapiens' 1.435508516404756;Eukaryota 0.5964291097600983;Simiiformes $ jq -r '.[].tax_totals|100*.identified/.total' SRR10903402.stat 71.32440955587016 $ seqkit grep -srp '^N*$' SRR10903402_1.fastq.gz|seqkit head -n1 @SRR10903402.1 1 length=151 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN + !!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!!! $ seqkit seq -s SRR10903402_[12].fastq.gz|grep '^N*$'|wc -l 177770 $ seqkit seq -s SRR10903402_[12].fastq.gz|wc -l 1353388
Human reads may have been masked for ethical reasons, since the SRA's website says: "Human metagenomic studies may contain human sequences and require that the donor provide consent to archive their data in an unprotected database. If you would like to archive human metagenomic sequences in the public SRA database please contact the SRA and we will screen and remove human sequence contaminants from your submission." [https://www.ncbi.nlm.nih.gov/sra/docs/submit/]
The NCBI has a tool called sra-human-scrubber which is
used to mask human reads with N bases in SRA submissions:
[https://ncbiinsights.ncbi.nlm.nih.gov/2023/02/02/scrubbing-human-sequences-sra-submissions/]
The Human Read Removal Tool (HRRT; also known as the Human Scrubber) is available on GitHub and DockerHub. The HRRT is based on the SRA Taxonomy Analysis Tool (STAT) that will take as input a fastq file and produce as output a fastq.clean file in which all reads identified as potentially of human origin are masked with 'N'.
You may also request that NCBI applies the HRRT to all SRA data linked to your submitted BioProject (more information below). When requested, all data previously submitted to the BioProject will be queued for scrubbing, and any future data submitted to the BioProject will be automatically scrubbed at load time.
This tool can be particularly helpful when a submission could be contaminated with human reads not consented for public display. Clinical pathogen and human metagenome samples are common submission types that benefit from applying the Human Scrubber tool.
For more information on genome data sharing policies, consult with institutional review boards and the NIH Genomic Data Sharing Policy. It is the responsibility of submitting parties to ensure that they have appropriate consent for human sequence data to be distributed publicly without access controls.
The usage message of the Human Scrubber tool also says that human reads are masked with N bases by default. [https://github.com/ncbi/sra-human-scrubber/blob/master/scripts/scrub.sh]
Wu et al. wrote: "Of the 384,096 contigs assembled by Megahit [9], the longest (30,474 nucleotides (nt)) had a high abundance and was closely related to a bat SARS-like coronavirus (CoV) isolate—bat SL-CoVZC45 (GenBank accession number MG772933)—that had previously been sampled in China, with a nucleotide identity of 89.1% (Supplementary Tables 1, 2)." Some Lankatards think that because there were about 400,000 contigs, it means that MEGAHIT produced about 400,000 candidate genomes for the virus but one of them was arbitrarily selected to be the genome of SARS2, and Lankatards also don't understand why Wu et al. selected the longest contig.
However the longest contig happened to be the contig for SARS2 by accident, and when I have ran MEGAHIT on other metagenomic samples which have contained a coronavirus, I have sometimes gotten bacterial contigs which were longer than the longest coronavirus contig. For example here where I ran MEGAHIT on a sample of bat shit that contained the bat sarbecovirus RmYN02, my longest contig was a contig for E. coli which was about 50,000 bases long, and the 2nd-longest and 3rd-longest contigs also matched E coli., but the contig for RmYN02 was only the 4th-longest contig:
$ curl 'https://www.ebi.ac.uk/ena/portal/api/filereport?accession=SRR12432009&result=read_run&fields=fastq_ftp'|sed 1d|cut -f2|tr \; \\n|sed s,^,ftp://,|xargs wget [...] $ megahit -1 SRR12432009_1.fastq.gz -2 SRR12432009_2.fastq.gz -o megarmyn02 [...] $ x='qseqid bitscore pident qcovs stitle qlen slen qstart qend sstart send';seqkit sort -lr megarmyn02/final.contigs.fa|seqkit head -n5|blastn -db nt -remote -num_alignments=1 -outfmt "6 $x">blast [...] $ (tr \ \\t<<<$x;awk '!a[$1]++' blast)|sed 's/, complete genome//'|column -ts$'\t' qseqid bitscore pident qcovs stitle qlen slen qstart qend sstart send k141_53690 92112 99.968 100 Escherichia coli strain Ec-RL2-1X chromosome 49929 4705802 1 49929 475668 425742 k141_64167 86149 100.000 100 Escherichia coli strain B chromosome 46651 4611841 1 46651 3954621 4001271 k141_22284 66979 100.000 100 Escherichia coli strain B chromosome 36270 4611841 1 36270 3501999 3465730 k141_29207 52181 99.996 100 Escherichia coli strain B chromosome 28260 4611841 1 28260 811981 840240
If Wu et al. would've had a larger number of reads, they might have also gotten bacterial contigs with a length over 30,474 bases, because then their additional reads might have filled in the gap between two contigs for different regions of a bacterial genome so that they would've been joined together into a single contig.
You can find out which contigs are viral by doing a BLAST search for each contig, like what Wu et al. did in their Supplementary Table 1. It shows that apart from the contig for SARS2, all of their other most abundant contigs were bacterial: [https://static-content.springer.com/esm/art%3A10.1038%2Fs41586-020-2008-3/MediaObjects/41586_2020_2008_MOESM1_ESM.pdf]

You can also find out which contigs are viral by trying to align all contigs against a FASTA file for virus reference sequences. For example here I ran MEGAHIT on Wu et al.'s raw reads. I got only 29,463 instead of 384,096 contigs because human reads were masked with N bases in the version of Wu et al.'s reads that were uploaded to the SRA:
$ brew install megahit -s # `-s` compiles from source because the bottle was compiled against GCC 9 so it gave me the error `dyld: Library not loaded: /usr/local/opt/gcc/lib/gcc/9/libgomp.1.dylib` [...] $ curl -s 'https://www.ebi.ac.uk/ena/portal/api/filereport?accession=SRR10971381&result=read_run&fields=fastq_ftp&format=tsv&download=true&limit=0'|sed 1d|cut -f2|tr \; \\n|sed s,^,ftp:,,|xargs wget -q $ megahit -1 SRR10971381_1.fastq.gz -2 SRR10971381_2.fastq.gz -o megawu [...] 2023-09-17 20:46:46 - 29463 contigs, total 14438186 bp, min 200 bp, max 29802 bp, avg 490 bp, N50 458 bp 2023-09-17 20:46:59 - ALL DONE. Time elapsed: 4545.123885 seconds
When I aligned all contigs against virus refseqs, I got only 8 aligned contigs. There was only contig for SARS2 because the contig covered almost the entire genome of the virus apart from a part of the 3' UTR so the genome was not split into multiple shorter contigs. There's 4 contigs which matched different parts of the human endogenous retrovirus K113, which is usually matched by human reads because the genomes of HERVs are incorporated as part of the human genome. There was also one short contig that matched a Streptococcus bacteriophage, and there were two short contigs which matched a parvovirus:
$ conda install -c bioconda bbmap
[...]
$ wget -q https://ftp.ncbi.nlm.nih.gov/refseq/release/viral/viral.1.1.genomic.fna.gz
$ seqkit fx2tab viral.1.1.genomic.fna.gz|sed $'s/A*\t$//'|seqkit tab2fx|bbmask.sh window=20 in=stdin out=viral.fa # bbmask masks low-complexity regions to avoid spurious matches
$ bowtie2-build --thread 3 viral.fa{}
[...]
$ bowtie2 -p3 --no-unal -x viral.fa -fU megawu/final.contigs.fa|samtools sort -@2 ->megawu.bam
$ x=megawu.bam;samtools coverage $x|awk \$4|cut -f1,3-6|(gsed -u '1s/$/\terr%\tname/;q';sort -rnk4|awk -F\\t -v OFS=\\t 'NR==FNR{a[$1]=$2;next}{print$0,sprintf("%.2f",a[$1])}' <(samtools view $x|awk -F\\t '{x=$3;n[x]++;len[x]+=length($10);sub(/.*NM:i:/,"");mis[x]+=$1}END{for(i in n)print i"\t"100*mis[i]/len[i]}') -|awk -F\\t 'NR==FNR{a[$1]=$2;next}{print$0"\t"a[$1]}' <(seqkit seq -n viral.fa|gsed 's/ /\t/;s/,.*//') -)|column -ts$'\t'
#rname endpos numreads covbases coverage err% name
NC_045512.2 29870 1 29801 99.769 0.01 Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1
NC_022518.1 9471 4 3435 36.2686 10.16 Human endogenous retrovirus K113 complete genome
NC_012756.1 31276 1 703 2.24773 4.69 Streptococcus phage PH10
NC_022089.1 3779 2 607 16.0625 3.29 Parvovirus NIH-CQV putative 15-kDa protein
The results above match the STAT results of the raw reads, which also include a handful of reads that are classified under parvoviruses and Pseudonomas phages.
MEGAHIT uses a default minimum length of 200 bases for contigs, but I only got two 200-base contigs. When I did a BLAST search for the first contig, it matched the human genome (because a small number of human reads remained in the raw reads at the SRA even though the vast majority of human reads were masked). And when I did a BLAST search for the other contig, it had a 100% match with about 80% coverage for ribosomal RNA sequences of Salmonella, Klebsiella, and some other bacteria:
$ seqkit seq -M 200 megawu/final.contigs.fa|seqkit seq -w0 >k141_7662 flag=0 multi=1.0000 len=200 GCAAGAGTATATTTGCCTAGCCTTGAGGATTTCGTTGGAAACGGGATTGTCTTCAGAGAA AATCTAGACAGAAGCATTCTCAGAAACTTCTTTGGGATGTTTGCATTCAAGTCACAGAGT AGAACATTCCCTTTGGTAGAGCAGGTTTGAAACACTCTTTTTTTAGTATATGGAAGTGGA CATTTGGAGCGCTTTCAGGC >k141_2082 flag=0 multi=1.0000 len=200 GGAAAGGGCGACGACGTACTTTCGGGTACGAGCATACACAGGTGCTGCATGGCTGTCGTC AGCTCGTGTTGTGAAATGTTGGGTTAAGTCCCGCAACGAGCGCAACCCTTATCCTTTGTT GCCAGCGGTCCGGCCGGGAACTCAAAGGAGACTGCCAGTGATAAACTGGAGGAAGGTGGG GATGACGTCAAGTCATCATG
There were two contigs that were exactly 1,000 bases long. The other contig matched Prevotella veroralis on BLAST, but there was no significant similarity found for the other contig:
$ seqkit seq -gM 1000 -m 1000 megawu/final.contigs.fa|seqkit seq -gw0 >k141_23439 flag=1 multi=15.0000 len=1000 GTGATGATACAAGGAACAGCAGAGAGCCGCATGGACGAGTCGAGCATGAATAATATCTATGTTCGCACAACTGCGGGCATGGCTCCAGTAAGTGAGTTCTGTACGTTGAAGCGTGTTTATGGACCGTCAAATATCACTCGTTTCAACCTCTTTACCTCTATTGCGGTTAATGCGACACCAGCAAACGGGGTCTCTTCTGGAGAGGCTATTAAGGCGGCAGAGGAAGTGGCAAAGCAAGTGTTACCACAGGGATATGGCTATGAGTTCTCTGGCTTGACACGTTCAGAGCAGGAGTCATCGAACTCAACAGCGATGATCTTCGTCCTTTGTATCGTGTTTGTTTACTTGATTCTTAGTGCACAGTATGAGAGTTATATCCTCCCATTAGCCGTTATCTTGTCAATACCATTCGGTCTTGCAGGTGCGTTCATATTCACGATGATCTTCGGACATAGCAACGATATCTACATGCAAATATCCCTGATTATGTTGATTGGATTGTTGGCAAAGAACGCCATCCTTATCGTTGAGTTTGCTCTCGAGCGTCGCCGAACAGGTATGGCAATCAAGTATGCAGCCATCTTAGGCGCTGGCGCACGTCTTCGTCCTATCCTTATGACGTCTTTGGCAATGGTTGTCGGATTGTTGCCATTGATGTTTGCAAGCGGTGTAGGTCATAATGGTAACCAGACATTGGGTGCTGCTGCCGTCGGAGGTATGTTGATTGGTACACTCTGTCAGGTGTTTGTTGTACCAGCTCTATTCGCTGGCTTCGAGTACTTACAAGAGCGTATTAAGCCTATCGAGTTTGAAGATGAGGCCAATCGCGACGTCATTAAGGAACTCGAGCAATACGCGAAAGGACCTGCACATGATTATAAGGTAGAAGAGTAGTACTATTTTAAAAGAACAATAATGAAGAATAAAAATATAATCATCATAGTCCTTGCAGCCCTGTCATTGACAGGCTGTAAGAGTCTCTATGGCAATTACGAGTTCT >k141_27896 flag=1 multi=8.0000 len=1000 TAAAGTTTCTCCGCATGATAAAAAAGCCTGGATTTAATGTTTTTGGAGTTATTAAAGAATTTAACAAAAAAGAAGATAAGGCGGCATTTGTTCCACCTGAATCAACAGTAAAAAAGTCTGAAACAAAGGTAGAAGACAAGAAAACTACGTAACCAAATAGATTCCAATAATGGAAGATTTAATAGGGGTTTTTAGTCGTTTGACCGGACAAAACGAGGATACTGTCAAGGAAGTATTAAAAGACCTTGAAGGCGAGAAATTTGCCAAAGCAGCTGAAAAGCTATTGAATGACAATTTCGGTAAAAAGTTAGCCGAAACAGAATCTAAAGCACGGATTGACGCAGAAAAGAAGCAGCGAAGCCTTATCCTTACTGAACAGGAAAAGAAACTAAAATCAAAGTACAGTATCGAATCATTCGATGACTTTGAAGACTTAGTAGAAAAGATCCACGAGTCAGGAAAGTTAAGTTCCGGTGCAGATGAAGTAAGCGTTAAAAAGATCGAGAAGCTCGAAAAGGACATTGAGAAGATAAAGCAATCTGAACAGACTATTAAAAGCGAATACGAAGCATTTAAAAGTACAACGGAACGCAATGTTTTATCAGGTTCAATCAGAAAGAGCGCACTTGAGTTTTTAGAGCAAAACAATTTCGTGGTAAGTGGTAAGCAAAGGGTAAATGATAATTTCATTGATGAGCTTTTGAATATCGCAGATTATGAGGTTGTGAACGGTGAATTTATTCCACTTATTAAAGGAACGAAAGAGAGGTTGATGGATGATGTAAGAAATCCTATATCCGCTCAAGACTTGTTTAAAAAGGTAGGAATGGATTACTACGATGTTAAAAATGGAAATGATAGAGAAACGGCAGGAGCGTTCAATACGAAGCCTACAGGAACAGGAGGTTCAAAATACAACATAAGTCAATCAAGACTTTAACAATGCAATAGCAGGGGCAAAGACTACAGAAGAACTTGATGTAATCGAAAAAGAATATCA
But basically many of these shorter contigs match different parts of bacterial genomes. And there's only a tiny number of contigs for viruses. And the original reads by Wu et al. also had a lot of human contigs because human reads had not been masked with N bases.
The sequencing runs at the SRA have been analyzed with a program called STAT, which stands for SRA Taxonomic Analysis Tool. STAT relies on matches to 32-base k-mers to classify each read under some node in a taxonomical tree, where it's able to classify some reads on the species-level but other reads are only classified at higher taxonomical levels. [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8450716/]
You can see a visual breakdown of the STAT results from SRA's website by going to the "Analysis" tab and clicking "Show Krona View". [https://trace.ncbi.nlm.nih.gov/Traces/?view=run_browser&acc=SRR10971381&display=analysis] It shows that about 61% of reads are unidentified, about 33% of reads are classified under bacteria out of which about half are classified under Prevotella, about 5% of reads are classified under eukaryotes, and about 0.2% of reads are classified under viruses:

The 4 most abundant leaf nodes in the STAT tree are all different species of Prevotella, followed by SARS2 on the 5th place, and then followed by other species of bacteria, humans, and other hominins from misclassified human reads:
$ curl 'https://trace.ncbi.nlm.nih.gov/Traces/sra-db-be/run_taxonomy?cluster_name=public&acc=SRR10971381'>SRR10971381.stat $ jq -r '[.[]|.tax_table[]|.parent]as$par|[.[]|.tax_table[]|select(.tax_id as$x|$par|index($x)|not)]|sort_by(-.total_count)[]|((.total_count|tostring)+";"+.org)' SRR10971381.stat|head -n30 171042;Prevotella salivae F0493 (Prevotella are common in the human oral cavity) 155602;Prevotella veroralis F0319 89078;Prevotella scopos JCM 17725 57886;Prevotella melaninogenica D18 54238;Severe acute respiratory syndrome coronavirus 2 51814;Leptotrichia sp. oral taxon 212 (Leptotrichia is a common genus of bacteria in human oral cavity) 41272;Prevotella nanceiensis DSM 19126 = JCM 15639 33716;Homo sapiens 28813;Prevotella veroralis DSM 19559 = JCM 6290 23601;Leptotrichia hongkongensis 14350;Prevotella pallens 12403;Prevotella sp. F0091 9451;Prevotella sp. C561 9261;Prevotella jejuni 8531;Prevotella histicola JCM 15637 = DNF00424 7944;Prevotella sp. oral taxon 313 7612;Prevotella sp. oral taxon 299 str. F0039 7356;Capnocytophaga gingivalis ATCC 33624 (common oral bacteria) 6631;Pan troglodytes 6391;Prevotella loescheii DSM 19665 = JCM 12249 = ATCC 15930 6229;Klebsiella pneumoniae (found in normal flora of the mouth, skin, and intestines) 6104;Veillonella parvula ATCC 17745 (normal part of oral flora) 5389;Prevotella sp. oral taxon 472 str. F0295 5251;Pongo abelii (Sumatran orangutan; misclassified human reads) 5059;Clostridioides difficile (intestinal bacteria which cause of diarrhea and colitis) 4810;Selenomonas sputigena ATCC 35185 (bacteria found in the upper respiratory tract) 4492;Acinetobacter baumannii (common in hospital-derived infections; almost exclusively isolated from hospital environments) 4020;Porphyromonas sp. oral taxon 278 str. W7784 3793;Gorilla gorilla gorilla 3791;Prevotella sp. ICM33
Almost all viral leaf nodes were classified under SARS2, but there were also about 700 reads classified under a CRESS virus which might be some kind of spurious matches, and there were a few reads for a parvovirus and a Pseudonomas phage (which both also got short contigs in my MEGAHIT run):
$ jq -r '.[]|.tax_table[]|[.org,.total_count,.tax_id,.parent]|@tsv' SRR10971381.stat|awk -F\\t '{out[$3]=$0;parent[$3]=$4;name[$3]=$1;nonleaf[$4]}END{for(i in out){j=parent[i];chain="";do{chain=(chain?chain":":"")name[j];j=parent[j]}while(j!="");print out[i]"\t"!(i in nonleaf)"\t"chain}}'|sort -nr|grep Virus|sort -t$'\t' -rnk2|awk -F\\t '$5{print$2 FS$1" - "$6}'|head -n20|column -ts$'\t'
54238 Severe acute respiratory syndrome coronavirus 2 - Severe acute respiratory syndrome-related coronavirus:Sarbecovirus:Betacoronavirus:Orthocoronavirinae:Coronaviridae:Cornidovirineae:Nidovirales:Riboviria:Viruses
693 CRESS virus sp. - CRESS viruses:unclassified ssDNA viruses:unclassified DNA viruses:unclassified viruses:Viruses
159 uncultured virus - environmental samples:Viruses
20 Parvovirus NIH-CQV - unclassified Parvovirinae:Parvovirinae:Parvoviridae:Viruses
15 Curvibacter phage P26059B - unclassified Autographivirinae:Autographivirinae:Podoviridae:Caudovirales:Viruses
13 Pseudomonas phage HU1 - unclassified Podoviridae:Podoviridae:Caudovirales:Viruses
12 Bat coronavirus RaTG13 - Severe acute respiratory syndrome-related coronavirus:Sarbecovirus:Betacoronavirus:Orthocoronavirinae:Coronaviridae:Cornidovirineae:Nidovirales:Riboviria:Viruses
9 Iodobacter phage PhiPLPE - Iodobacter virus PLPE:Iodovirus:Myoviridae:Caudovirales:Viruses
9 Bat SARS-like coronavirus - Severe acute respiratory syndrome-related coronavirus:Sarbecovirus:Betacoronavirus:Orthocoronavirinae:Coronaviridae:Cornidovirineae:Nidovirales:Riboviria:Viruses
8 Rhodoferax phage P26218 - unclassified Podoviridae:Podoviridae:Caudovirales:Viruses
8 Prokaryotic dsDNA virus sp. - unclassified dsDNA viruses:unclassified DNA viruses:unclassified viruses:Viruses
8 Pangolin coronavirus - unclassified Betacoronavirus:Betacoronavirus:Orthocoronavirinae:Coronaviridae:Cornidovirineae:Nidovirales:Riboviria:Viruses
5 uncultured cyanophage - environmental samples:unclassified bacterial viruses:unclassified viruses:Viruses
5 Rimavirus - Siphoviridae:Caudovirales:Viruses
4 Genomoviridae sp. - unclassified Genomoviridae:Genomoviridae:Viruses
3 Streptococcus phage Javan363 - unclassified Siphoviridae:Siphoviridae:Caudovirales:Viruses
3 Sewage-associated circular DNA virus-14 - unclassified viruses:Viruses
3 RDHV-like virus SF1 - unclassified viruses:Viruses
3 Paramecium bursaria Chlorella virus NW665.2 - unclassified Chlorovirus:Chlorovirus:Phycodnaviridae:Viruses
3 Microviridae sp. - unclassified Microviridae:Microviridae:Viruses
Trinity is a lot slower than MEGAHIT, and it requires more RAM and
more free disk space, so the first few times when I ran it on my local
computer, I either ran out of disk space, or I ran out of RAM so it had
to use swap and I terminated the run. I think it required around 20-30
GB RAM and around 50 GB disk space to assemble Wu et al.'s reads. The
--max_memory flag which specifies a maximum limit for RAM
and swap use cannot be omitted, and one of the first times I ran Trinity
I think it was interrupted in the middle of the run because it exceeded
the --max_memory limit. Running Trinity took about 24 hours
on a server with 16 cores:
trinity --seqType fq --left SRR10971381_1.fastq --right SRR10971381_2.fastq --max_memory 50G --CPU 16 --output trinityout
The contigs produced by Trinity can be downloaded from here: f/Trinity.fasta.gz.
I got a total of only 159,532 contigs even though Wu et al. wrote that they got 1,329,960 contigs with Trinity, but it's probably because I used the reads that are publicly available at the SRA where almost all human reads are masked, so I got only a small number of human contigs.
My Trinity run produced six contigs for SARS2 but the longest contig is 29,869 bases even though Wu et al.'s longest contig was only 11,760 bases. It might be because I didn't trim the reads before I ran Trinity, or because human reads were masked in my run but not in Wu et al.'s run, or because I used a different version of Trinity than Wu et al.:
$ wget https://sars2.net/f/Trinity.fasta.gz
$ seqkit stat Trinity.fasta.gz
file format type num_seqs sum_len min_len avg_len max_len
Trinity.fasta.gz FASTA DNA 159,532 46,995,710 186 294.6 29,879
$ curl 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=MN908947.3'>sars2.fa
$ bowtie2-build sars2.fa{,}
[...]
$ bowtie2 -p4 --no-unal -x sars2.fa -fU Trinity.fasta.gz|samtools sort ->trinity.bam
159532 reads; of these:
159532 (100.00%) were unpaired; of these:
159526 (100.00%) aligned 0 times
6 (0.00%) aligned exactly 1 time
0 (0.00%) aligned >1 times
0.00% overall alignment rate
$ samtools view trinity.bam|ruby -ane'l=$F[9].length;s=$F[3].to_i;e=s+l-$F[5].scan(/\d+(?=I)/).map(&:to_i).sum-1;puts [$F[0],s,e,l,$F.grep(/NM:i/)[0][/\d+/],$F[5]]*" "'|(echo contig_name start end length mismatches cigar_string;cat)|column -t
contig_name start end length mismatches cigar_string
TRINITY_DN1442_c18_g1_i2 1 29874 29879 10 4M5I29870M
TRINITY_DN1442_c18_g1_i4 77 26478 26402 4 21432M1D4970M
TRINITY_DN131_c1_g1_i1 19825 20128 306 17 299M2I5M
TRINITY_DN1442_c18_g1_i7 21531 26479 4998 54 7M5I3M5I3M4I8M35I4928M
TRINITY_DN1442_c18_g1_i1 26425 29871 3476 41 4M2I10M2D2M1I5M5I2M11I7M1D5M8I4M2I3408M
TRINITY_DN8024_c1_g1_i1 28394 28626 233 9 233M
If my longest contig is compared to the third version of Wuhan-Hu-1, the only differences are that my contig has 5 extra nucleotides at the start, one nucleotide change in the middle, and it's missing the last 29 bases of the poly(A) tail:
$ R -e 'if(!require("BiocManager"))install.packages("BiocManager");BiocManager::install("Biostrings")'
[...]
$ aldi2()(cat "$@">/tmp/aldi;Rscript --no-init-file -e 'm=t(as.matrix(Biostrings::readDNAStringSet("/tmp/aldi")));diff=which(rowSums(m!=m[,1])>0);split(diff,cumsum(diff(c(-Inf,diff))!=1))|>sapply(\(x)c(if(length(x)==1)x else paste0(x[1],"-",tail(x,1)),apply(m[x,,drop=F],2,paste,collapse="")))|>t()|>write.table(quote=F,sep=";",row.names=F)')
$ seqkit sort -lr Trinity.fasta.gz|seqkit head -n1|cat - sars2.fa|mafft --thread 4 -|aldi2
[...]
;TRINITY_DN1442_c18_g1_i2 len=29879 path=[0:0-79 1:80-80 2:81-21515 4:21516-21544 5:21545-21565 6:21566-21568 7:21569-21569 8:21570-26483 9:26484-29878];NC_045512.2 Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1, complete genome
1-5;CGGGG;-----
21666;G;A
29880-29908;-----------------------------;AAAAAAAAAAAAAAAAAAAAAAAAAAAAA
The reason why the longest contig is shown to have 10 mismatches in the BAM file is that Bowtie2 doesn't allow gaps within the first 4 bases of the read by default, so it treated the first 4 bases as mismatches and it added a 5-base insert after them:
$ bowtie2 -h|grep gbar --gbar <int> disallow gaps within <int> nucs of read extremes (4)
I don't know why Trinity produced 6 different contigs without merging them into a single contig, even though the longest contig covered the entire genome apart from the poly(A) tail. However it could be because the reads were not trimmed properly so there were mismatches in the middle of the contigs.
Out of my 6 contigs, the shortest 233-base contig matched positions 28,394 to 28,626 of Wuhan-Hu-1. It has 3 wrong bases at the start of the contig, but it might be because many reads have 3-5 bases of extra crap at the ends but I didn't trim the ends of reads. The shortest contig also has 6 errors where a T base was changed to an A base, which may have been because A bases have the lowest average quality and I didn't trim low-quality bases from the ends of reads:
$ seqkit grep -p TRINITY_DN8024_c1_g1_i1 Trinity.fasta.gz|seqkit seq -rp|cat - sars2.fa|mafft --thread 4 --clustalout -
[...]
TRINITY_DN8024_ -------------gtaccccaaggtttacccaataatactgcgtcttggttcaccgctct
NC_045512.2 atcaaaacaacgtcggccccaaggtttacccaataatactgcgtcttggttcaccgctct
.********************************************
TRINITY_DN8024_ cacacaacatggcaaggaagaccttaaattccctcgaggacaaggcgttccaattaacac
NC_045512.2 cactcaacatggcaaggaagaccttaaattccctcgaggacaaggcgttccaattaacac
*** ********************************************************
TRINITY_DN8024_ caaaagcagtccagatgaccaaataggctactaccgaagagctaccagacgaaatcgtgg
NC_045512.2 caatagcagtccagatgaccaaattggctactaccgaagagctaccagacgaattcgtgg
*** ******************** **************************** ******
TRINITY_DN8024_ tggtgacggaaaaatgaaagatctcagtccaagaaggtatttctactacctaggaactgg
NC_045512.2 tggtgacggtaaaatgaaagatctcagtccaagatggtatttctactacctaggaactgg
********* ************************ *************************
TRINITY_DN8024_ gccaga------------------------------------------------------
NC_045512.2 gccagaagctggacttccctatggtgctaacaaagacggcatcatatgggttgcaactga
******
[...]
When I ran Trinity a second time so that I trimmed the reads with
fastp, I got 109,806 contigs with a maximum length of
31,241. Now only two contigs aligned against SARS2, but the longer
contig was 31,241 bases long because it had a 1,379-base insertion in
the middle, and the shorter contig also had the same 1,379-base
insertion in the middle:
$ fastp -f 5 -t 5 -53 -i SRR10971381_1.fastq.gz -I SRR10971381_2.fastq.gz -o SRR10971381_1.trim.fq -O SRR10971381_2.trim.fq [...] $ trinity --seqType fq --left SRR10971381_1.trim.fq --right SRR10971381_2.trim.fq --max_memory 30G --CPU 16 --output trinityout2 [...] $ bowtie2 -p4 --no-unal -x sars2.fa -fU Downloads/trinityout2.Trinity.fasta.gz|samtools sort ->trinity2.bam [...] $ samtools view trinity2.bam|ruby -ane'l=$F[9].length;s=$F[3].to_i;e=s+l-$F[5].scan(/\d+(?=I)/).map(&:to_i).sum-1;puts [$F[0],s,e,l,$F.grep(/NM:i/)[0][/\d+/],$F[5]]*" "'|(echo contig_name start end length mismatches cigar_string;cat)|column -t contig_name start end length mismatches cigar_string TRINITY_DN39043_c0_g1_i2 4 29865 31241 1379 27790M1379I2072M TRINITY_DN39043_c0_g1_i1 21487 29852 9755 1413 6M4D5M1I3M3I2M3I28M1I2M2I6M9D6242M1379I2072M
When I did a BLAST search for the insert, it started with a 64-base segment which was identical to bases 29,109-29,172 of Wuhan-Hu-1 which was followed by a 1,315-base segment which was identical to bases 27,794-29,108 of Wuhan-Hu-1. So even though the starting position of the first segment was one base bigger than the ending position of the second segment, for some reason the order of the two segments was flipped in the insert.
Mark Bailey wrote: "As has been noted, 'bat SL-CoVZC45' was an in silico genome, 29,802 nucleotides in length, invented in 2018, that was used by Fan Wu et al. as a template genome for the invention of the SARS-CoV-2 genome." [https://drsambailey.substack.com/p/a-farewell-to-virology-expert-edition]
His misconception is based on a paragraph of the paper by Wu et al. said: "The viral genome organization of WHCV was determined by sequence alignment to two representative members of the genus Betacoronavirus: a coronavirus associated with humans (SARS-CoV Tor2, GenBank accession number AY274119) and a coronavirus associated with bats (bat SL-CoVZC45, GenBank accession number MG772933). The un-translational regions and open-reading frame (ORF) of WHCV were mapped on the basis of this sequence alignment and ORF prediction. The WHCV viral genome was similar to these two coronaviruses (Fig. (Fig.11 and Supplementary Table 3). The order of genes (5' to 3') was as follows: replicase ORF1ab, spike (S), envelope (E), membrane (M) and nucleocapsid (N)." [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7094943/]
However the authors meant that after they had already assembled the complete genome of SARS2, they aligned it against the genomes of SARS1 and ZC45 in order to annotate the proteins corresponding to each ORF. They didn't mean that they used SARS1 as a reference genome when they assembled the raw reads.
You can try running NCBI's ORFfinder on the genome of SARS2. Go here,
enter NC_045512 as the accession number, enable the option
to ignore nested ORFs, and click submit:
https://www.ncbi.nlm.nih.gov/orffinder/. It shows
that the third ORF on the positive strand starts at position 21,536 and
ends at position 25,384:
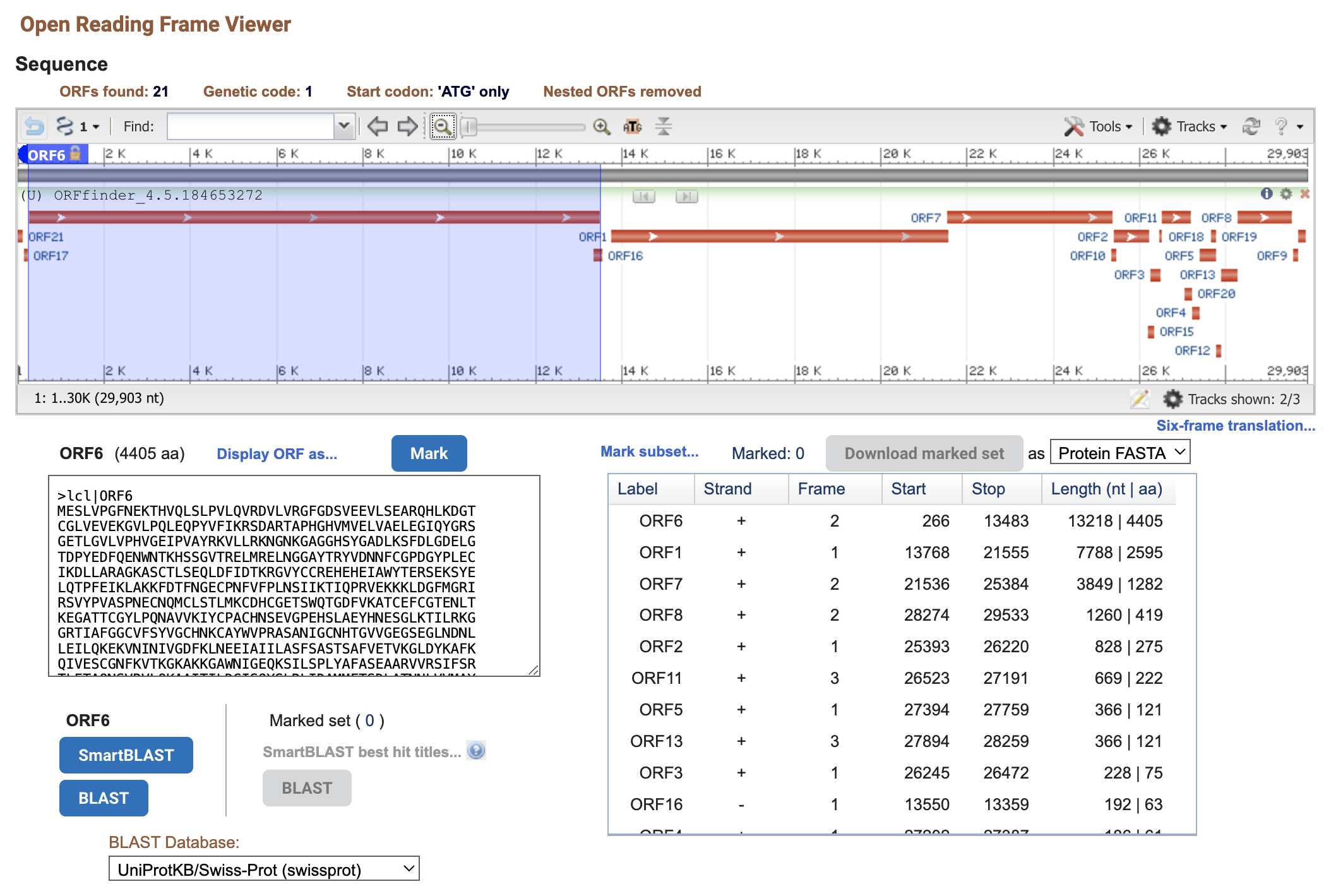
In order to know what name you should give to the protein coded by the ORF, you can check which protein SARS1 has around the same region. But it happens to be the spike protein:
$ curl -s 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta_cds_aa&id=NC_004718'|grep \>|sed 's/.*protein=//;s/\].*location=/: /;s/\].*//' ORF1ab polyprotein: join(265..13392,13392..21485) ORF1a polyprotein: 265..13413 spike glycoprotein: 21492..25259 ORF3a protein: 25268..26092 ORF3b protein: 25689..26153 small envelope protein: 26117..26347 membrane glycoprotein M: 26398..27063 ORF6 protein: 27074..27265 ORF7a protein: 27273..27641 ORF7b protein: 27638..27772 ORF8a protein: 27779..27898 ORF8b protein: 27864..28118 nucleocapsid protein: 28120..29388 ORF9b protein: 28130..28426 ORF9a protein: 28583..28795
But for example the ORF6 of Wuhan-Hu-1 starts at position 27,202, so if you simply looked at which ORF in SARS1 is the closest to it but you didn't align the sequences, it would be ORF7a. But if you align the sequences, then the ORF6 of SARS2 and SARS1 will have the same starting position so it's easier to do the annotation:
$ curl 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=NC_004718,NC_045512'>sars12.fa $ mafft --quiet sars12.fa>sars12.aln $ seqkit head -n1 sars12.fa|seqkit subseq -r27074:27100|seqkit locate -if- sars12.aln|sed 1d|cut -f5,6|seqkit subseq -r $(tr \\t :) sars12.aln >NC_004718.3 SARS coronavirus Tor2, complete genome atgtttcatcttgttgacttccaggtt >NC_045512.2 Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1, complete genome atgtttcatctcgttgactttcaggtt
The website of the NCBI's sequence read archive has a tab which shows a breakdown of what percentage of reads are estimated to come from different organisms and different taxonomial nodes. The tab displays the results a program called SRA Taxonomy Analysis Tool (STAT), which searches for 32-base k-mers among the reads which match a set of reference sequences. A database which contains the number of reads which match each taxonomical node for each SRA run is currently about 500 GB in size, but you can search it through BigQuery at the Google Cloud Platform. [https://www.ncbi.nlm.nih.gov/sra/docs/sra-cloud-based-taxonomy-analysis-table/] I tried using BigQuery to search for the oldest sequencing runs which matched over 10,000 reads of SARS2 (taxid2697049):
select * from `nih-sra-datastore.sra.metadata` as m, `nih-sra-datastore.sra_tax_analysis_tool.tax_analysis` as tax where m.acc=tax.acc and tax_id=2697049 and total_count>10000 order by releasedate
I got 17 results from before 2020 but all of them had zero reads which aligned against Wuhan-Hu-1 when I used Bowtie2 with the default settings. The oldest results I found which had reads that aligned against SARS2 were runs for the samples WHU01 and WHU02 which were submitted by Wuhan University and published on January 18th 2020 (in an unknown timezone). [https://www.ncbi.nlm.nih.gov/sra/?term=SRR10903402] When I ran MEGAHIT on the reads of the WHU01 sample that I didn't do any trimming, my longest contig was 17,699 bases long, and it was identical to positions 12197-29895 of Wuhan-Hu-1:
wget ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR109/002/SRR10903402/SRR10903402_{1,2}.fastq.gz # download from ENA because downloading from SRA is too slow
curl 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=MN908947.3'>sars2.fa
megahit -1 SRR10903402_1.fastq.gz -2 SRR10903402_2.fastq.gz -o megawuhanjan
seqkit sort -lr megawuhanjan/final.contigs.fa|seqkit head -n1|cat - sars2.fa|mafft --clustalout -
When I trimmed the reads with fastp so that I used the
default settings apart from a longer minimum length, almost half of the
read pairs got an adapter sequence trimmed, so the adapter sequences may
have earlier prevented assembling a longer contig. Now my longest contig
was 29,809 bases long, and it missed the first 76 bases and last 19
bases of Wuhan-Hu-1 (which should demonstrate that it's common for
MEGAHIT contigs to miss pieces from the start or end of the genome):
$ fastp -i SRR10903402_1.fastq.gz -I SRR10903402_2.fastq.gz -o SRR10903402_1.trim.fq.gz -O SRR10903402_2.trim.fq.gz
Read1 before filtering:
total reads: 676694
total bases: 101889418
[...]
Filtering result:
reads passed filter: 1126442
reads failed due to low quality: 193854
reads failed due to too many N: 0
reads failed due to too short: 33092
reads with adapter trimmed: 287572
bases trimmed due to adapters: 12910527
[...]
$ megahit -1 SRR10903402_1.trim.fq.gz -2 SRR10903402_2.trim.fq.gz -o megawuhanjan3
[...]
2023-05-20 14:22:39 - 425 contigs, total 246369 bp, min 286 bp, max 29809 bp, avg 579 bp, N50 540 bp
2023-05-20 14:22:39 - ALL DONE. Time elapsed: 72.962219 seconds
$ seqkit sort -lr megawuhanjan3/final.contigs.fa|seqkit head -n1|cat sars2.fa -|mafft ->temp.aln
[...]
$ Rscript --no-init-file -e 'm=t(as.matrix(Biostrings::readDNAStringSet("temp.aln")));pick=which(rowSums(m!=m[,1])>0);writeLines(unlist(lapply(split(pick,cummax(c(1,diff(pick)))),\(x)paste(x[1],tail(x,1),paste(apply(m[x,],2,paste,collapse=""),collapse=" ")))))'
1 76 ATTAAAGGTTTATACCTTCCCAGGTAACAAACCAACCAACTTTCGATCTCTTGTAGATCTGTTCTCTAAACGAACT ----------------------------------------------------------------------------
29885 29903 AAAAAAAAAAAAAAAAAAA T------------------
When I did variant calling for the reads submitted by Wuhan University so that I used Wuhan-Hu-1 as the reference, I didn't get a single variant as expected:
brew install bowtie2 samtools bcftools
bowtie2-build sars2.fa{}
x=wuhanjan;bowtie2 -p3 -x sars2.fa -1 SRR10903402_1.trim.fq.gz -2 SRR10903402_2.trim.fq.gz --no-unal|samtools sort -@2 ->$x.bam
samtools index $x.bam;bcftools mpileup -f sars2.fa $x.bam|bcftools call -mv -Ob -o $x.bcf;bcftools index $x.bcf;bcftools consensus -f sars2.fa $x.bcf>$x.fa
Some of the reads aligned by Bowtie2 had a longer poly(A) tail than
Wuhan-Hu-1, so I tried aligning the untrimmed reads against a version of
Wuhan-Hu-1 with an extended poly(A) tail. I found one read where the
poly(A) tail started 73 bases from the end of the read, even though it
had a bunch of non-A bases near the end of the read and it only had 48
contiguous A bases. But I also got another read with 47 contiguous A
bases, another with 39, and another with 38. And may of the long
segments of A bases are preceded by GAATGAC which are also
the last bases of Wuhan-Hu-1 before the poly(A) tail. So I don't know if
the included samples of SARS2 which actually had a poly(A) tail longer
than 33 bases or if its's just some kind of an artifact:
$ (cat sars2.fa;echo AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA)>sars2a.fa
$ bowtie2-build sars2a.fa{}
[...]
$ bowtie2 -x sars2a.fa -1 SRR10903402_1.fastq.gz -2 SRR10903402_2.fastq.gz --no-unal -p3|samtools sort -@2 ->sorted54.bam
[...]
$ samtools view sorted54.bam|cut -f4,10,17|tail -n8|column -t # show reads with highest starting position (NM is number of mismatches)
29755 ACGTGTGCTCTTCCGATCTGCTAGGGAGAGCTGCCTATATGGAAGAGCCCTAATTTGTAAAATTAATTTTAGTAGTGCTATCCCCATGTGATTTTAATAGCTTTTTAGGAGAATGACAAAAAAAAAAAAAAAAAAAAAAAAAAAGAAAAAA NM:i:14
29756 AGTGTACAGTGAACAATGCTAGGGAGAGCTGCCTATATGGAAGAGCCCTAATGTTTAAAATTAATTTTAGTAGTGCTATCCCCATGTGATTTTAATAGCTTTTTAGGAGAATGACAAAAAAAAAAAAAAAAAAAAAAAAAATCAAACACA NM:i:6
29757 GTATCAAGTAAAAAATGTTAGGGAGAATTGCCTAAATGGAAAAGCCATAATTTTTAAAAATAATTTTAGTAGTGTTATCCCCATGTAATTTTAAAAATTTCTTAGGAGAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAGAG NM:i:24
29759 GTACAGTGAACAATGCTAGGGAGAGCTGCCTATATGGAAGAGCCCTAATGTGTAAAATTAATTTTAGTAGTGCTATCCCCATGTGATTTTAAAAGCTTCTTAGGAGAATGACAAAAAAAAAAAAAACAAAAAAAAAAAATCAAACACAAA NM:i:6
29761 ACAGTGAACAATGCTAGGGAGAGCTGCCTATATGGAAGAGCCTTAATGTGTAAAATTAATGTTAGGAGTGCTATCTCCATGTGATTTTAATAGCTTCTTAGGAGAATGACAAAAAAAAAAAAAAAAAAAATGATATGAATAGCGTTGGTTA NM:i:19
29762 AAGGAAAAAAAGCTAGGAAGAGATGCCTAAATGGAAAAAACAAAATTTTAAAAATTAATTTTAGGGGGGGTATCCCCATGTGATTTTAATAATTTTTTAGGAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAGAA NM:i:28
29765 TGAACAATGCTAGGGGGAGCTGCCTATATGGAAAAGCCCTAATGTGTAAAATTAATTTTAGTAGTGCTATCCCCATGTGATTTTAAAAGCTTCTTAGGAGAAAGACAAAAAAAAAAAAAAAAAATAAAAAAAAAAAAAATCAAACAC NM:i:9
29773 GCTAGGGAGAGCTGCCTATATGGAAGAGCCCTAATGTGTAAAATTAATTTTAGTAGTGCTATCCCCATGTGATTTTAATAGCTTCTTAGGAGAATGACAAAAAAAAAAAAAAAAAAAAAAAAAAAAGAACGGAAGAGCGGCGGAGAGGAAA NM:i:15
29773 GCTAGGGAGAGCTGCCTATATGGAAGAGCCCTAATGTGTAAAATTAATTTTAGTAGTGCTATCCCCATGTGATTTTAATAGCTTCTTAGGAGAATGACAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAACGGAAAAGCAACGGA NM:i:8
29793 TGGAAGAGCCCTAATGAGTAAAATTAATTTTAGTAGTGCTATCCCCATGTGATTTTAATAGCTTCTTAGGAGAATGACAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAGGAAAAAGCAAGTGGAGGAAAAAAA NM:i:11
$ samtools view sorted54.bam|cut -f10|tail -n100|grep -Eo A+|awk '{print length}'|sort -n|tail # print the length of the longest consecutive segments of A bases among the last 100 reads
26
27
27
27
28
34
38
39
47
48
The last read shown above has 48 consecutive A bases. Its reverse pair has 47 consecutive T bases even though they're at a different spot so that there's only 4 bases before them, even though in the forward pair there's 25 bases after the segment of contiguous A bases:
$ seqkit grep -p $(samtools view sorted54.bam|tail -n1|cut -f1) SRR10903402_1.fastq.gz |seqkit seq -s TTGATTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTCCTAAAAAACTTTTAAAATCAATTGGGGTTAGCACTACTAAAATTAATTTTACAAATTTGGGCTTTTCCAAAATGGGAAAAACACACGTCTAAACCCCAG
Some people were saying that it was somehow problematic that the full poly(A) tail of Wuhan-Hu-1 could not be reproduced from the metagenomic sequencing run of Wu et al. 2020. However above I have shown that there's another early metagenomic sequencing run of SARS2 where the reads actually feature a long poly(A) tail.
When I additionally ran MEGAHIT on the reads of the WHU02 sample, my longest contig was 29,997 bases long, and it was otherwise identical to Wuhan-Hu-1 except it had an insertion of 109 bases at the beginning, there was one nucleotide change after the insertion, and the poly(A) tail was missing the last 15 bases. When I did a BLAST search for the 109-base insertion, I found that it was 100% identical to the reverse complement of the segment between positions 191 and 299 in Wuhan-Hu-1.
In order to test if the genome of SARS2 can be assembled from other
metagenomic datasets, I searched the SRA for the phrase
human bronchoalveolar pneumonia.
[https://www.ncbi.nlm.nih.gov/sra/?term=human+bronchoalveolar+pneumonia]
I picked the first result which was from a study titled "Bronchoalveolar Lavage Fluid Metagenomic Next-Generation
Sequencing in non-severe and severe Pneumonia".
[https://www.ncbi.nlm.nih.gov/sra/?term=SRR22183690]
When I ran MEGAHIT with the default settings, I got a total of about 500
contigs with a maximum length of about 8,000. When I aligned the contigs
against a file of about 15,000 virus reference sequences, I got only 14
aligned contigs, none of which matched SARS2:
$ wget -q ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR221/090/SRR22183690/SRR22183690_{1,2}.fastq.gz
$ brew install megahit -s # `-s` compiles from source because the bottle was compiled against GCC 9 so it gave me the error `dyld: Library not loaded: /usr/local/opt/gcc/lib/gcc/9/libgomp.1.dylib`
[...]
$ megahit -1 SRR22183690_1.fastq.gz -2 SRR22183690_2.fastq.gz -o megapneumonia
[...]
$ seqkit stat megapneumonia/final.contigs.fa
file format type num_seqs sum_len min_len avg_len max_len
megapneumonia/final.contigs.fa FASTA DNA 508 230,544 213 453.8 8,451
$ brew install bowtie2 seqkit gnu-sed
[...]
$ wget -q https://ftp.ncbi.nlm.nih.gov/refseq/release/viral/viral.1.1.genomic.fna.gz
$ seqkit fx2tab viral.1.1.genomic.fna.gz|sed $'s/A*\t$//'|seqkit tab2fx>viral.fa
$ bowtie2-build viral.fa{,}
[...]
$ bowtie2 -p3 -x viral.fa -fU megapneumonia/final.contigs.fa --no-unal|samtools sort ->megapneumonia.bam
[...]
$ x=megapneumonia.bam;samtools coverage $x|awk \$4|cut -f1,3-6|(gsed -u '1s/$/\terr%\tname/;q';sort -rnk4|awk -F\\t -v OFS=\\t 'NR==FNR{a[$1]=$2;next}{print$0,sprintf("%.2f",a[$1])}' <(samtools view $x|awk -F\\t '{x=$3;n[x]++;len[x]+=length($10);sub(/.*NM:i:/,"");mis[x]+=$1}END{for(i in n)print i"\t"100*mis[i]/len[i]}') -|awk -F\\t 'NR==FNR{a[$1]=$2;next}{print$0"\t"a[$1]}' <(seqkit seq -n viral.fa|gsed 's/ /\t/;s/, .*//') -)|column -ts$'\t'
#rname endpos numreads covbases coverage err% name
NC_022518.1 9472 7 2334 24.641 5.13 Human endogenous retrovirus K113 complete genome
NC_050152.1 101660 1 250 0.245918 0.00 Enterobacteria phage P7
NC_032111.1 163005 1 143 0.0877274 3.47 BeAn 58058 virus
NC_018464.1 927 1 87 9.38511 0.45 Shamonda virus N and NSs genes
NC_007610.1 131384 1 45 0.0342507 0.22 Listeria bacteriophage P100
NC_037667.1 2077288 1 36 0.00173303 0.00 Pandoravirus quercus
NC_021858.1 1908524 1 34 0.00178148 0.00 Pandoravirus dulcis
NC_011189.1 3418 1 34 0.994734 0.00 Mikania micrantha mosaic virus RNA2
The contigs which matched human endogenous retrovirus K113 probably just matched the human genome. When I did a BLAST search for the longest contig, it got a 98.9% identical match to both some human endogenous retroviruses and to human DNA. When I downloaded other metagenomic sequencing runs for human lung or BALF samples from SRA and aligned the reads against virus refseqs, human endogenous retrovirus K113 got the highest coverage for most runs, including SRR22183691, SRR22183705, SRR1553689, and SRR22183702. And when I did a BLAST search for the full sequence of K113, it was about 98.8% identical to a segment of human DNA.
Below I aligned 194 sequences of SARS1 and then printed the last 100 bases of the alignment. You can see that there's a lot of variation in the length of the poly(A) tail, and many sequences are even missing a part of the 3' end before the poly(A) tail:
$ wget https://sars2.net/f/sarbe.{pid,aln.xz}
$ awk -F\\t 'NR==1{for(i=2;i<=NF;i++)if($i~/Tor2/)break;next}$1!=x{print$i,$1}' sarbe.pid|awk '$1>=99{print$2}'|seqkit grep -f- sarbe.aln.xz|mafft --thread 4 --quiet ->sars1.aln
$ seqkit subseq -r-100:-1 sars1.aln|seqkit fx2tab|awk -F\\t '{print$2,$1}'|sed 's/, complete genome//'
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaa------------------ AY278489.2 SARS coronavirus GD01
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaa------------------------------- AY390556.1 SARS coronavirus GZ02
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttctta---------------------------------------------- AY304488.1 SARS coronavirus SZ16
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatcac----------------------------------- AY304486.1 SARS coronavirus SZ3
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- NC_004718.3 SARS coronavirus Tor2
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- DQ898174.1 SARS coronavirus strain CV7
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY323977.2 SARS coronavirus HSR 1
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- AY278741.1 SARS coronavirus Urbani
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- JN854286.1 SARS coronavirus HKU-39849 isolate recSARS-CoV HKU-39849
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- JQ316196.1 SARS coronavirus HKU-39849 isolate UOB
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- AY714217.1 SARS Coronavirus CDC#200301157
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaa--------------------------------- AY502932.1 SARS coronavirus TW9
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaa--------------------------------- AY502923.1 SARS coronavirus TW10
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaa--------------------------------- AY502931.1 SARS coronavirus TW8
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaa--------------------------------- AY502930.1 SARS coronavirus TW7
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaa--------------------------------- AY502929.1 SARS coronavirus TW6
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaa--------------------------------- AY502928.1 SARS coronavirus TW5
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaa--------------------------------- AY502927.1 SARS coronavirus TW4
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaa--------------------------------- AY502925.1 SARS coronavirus TW2
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaa--------------------------------- AY291451.1 SARS coronavirus TW1
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaa--------------------------------- AY502926.1 SARS coronavirus TW3
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- AP006561.1 SARS coronavirus TWY genomic RNA
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- AP006560.1 SARS coronavirus TWS genomic RNA
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- AP006559.1 SARS coronavirus TWK genomic RNA
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- AP006557.1 SARS coronavirus TWH genomic RNA
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- AY362698.1 SARS coronavirus TWC2
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- AY362699.1 SARS coronavirus TWC3
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaa------------------------------- EU371559.1 SARS coronavirus ZJ02
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- AY283798.2 SARS coronavirus Sin2774
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- AY283796.1 SARS coronavirus Sin2679
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- AY283794.1 SARS coronavirus Sin2500
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- AY427439.1 SARS coronavirus AS
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- AY283797.1 SARS coronavirus Sin2748
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaa---------------------- AY310120.1 SARS coronavirus FRA
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- AY291315.1 SARS coronavirus Frankfurt 1
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- AY283795.1 SARS coronavirus Sin2677
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- AY321118.1 SARS coronavirus TWC
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- DQ497008.1 SARS coronavirus strain MA-15
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- MK062180.1 SARS coronavirus Urbani isolate icSARS-MA
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- MK062179.1 SARS coronavirus Urbani isolate icSARS
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- AP006558.1 SARS coronavirus TWJ genomic RNA
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY282752.2 SARS coronavirus CUHK-Su10
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY345987.1 SARS coronavirus CUHK-AG02
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY345986.1 SARS coronavirus CUHK-AG01
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY394998.1 SARS coronavirus LC1
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY345988.1 SARS coronavirus CUHK-AG03
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY350750.1 SARS coronavirus PUMC01
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY278554.2 SARS coronavirus CUHK-W1
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY394993.1 SARS coronavirus HGZ8L2
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY394992.1 SARS coronavirus HZS2-C
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY394990.1 SARS coronavirus HZS2-E
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY394989.1 SARS coronavirus HZS2-D
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY394991.1 SARS coronavirus HZS2-Fc
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY394987.1 SARS coronavirus HZS2-Fb
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY394983.1 SARS coronavirus HSZ2-A
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY864805.1 SARS coronavirus BJ162
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY864806.1 SARS coronavirus BJ202
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY394982.1 SARS coronavirus HGZ8L1-B, partial genome
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- GU553365.1 SARS coronavirus HKU-39849 isolate TCVSP-HARROD-00003
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- GU553364.1 SARS coronavirus HKU-39849 isolate TCVSP-HARROD-00002
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- GU553363.1 SARS coronavirus HKU-39849 isolate TCVSP-HARROD-00001
---------------------------------------------------------------------------------------------------- AY348314.1 SARS coronavirus Taiwan TC3
---------------------------------------------------------------------------------------------------- AY338174.1 SARS coronavirus Taiwan TC1
---------------------------------------------------------------------------------------------------- AY338175.1 SARS coronavirus Taiwan TC2
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgaca---------------------------------- AY559096.1 SARS coronavirus Sin850
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaa--------------------------------- AY559095.1 SARS coronavirus Sin847
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaa--------------------------------- AY559085.1 SARS coronavirus Sin848
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgaca---------------------------------- AY559093.1 SARS coronavirus Sin845
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaa------------------------------ AY559087.1 SARS coronavirus Sin3725V
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaa------------------------ AY559084.1 SARS coronavirus Sin3765V
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatga------------------------------------ AY559086.1 SARS coronavirus Sin849
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgaca---------------------------------- AY559081.1 SARS coronavirus Sin842
gtgt-aaaattaattttagtagtgctatccccatgtga-------------------------------------------------------------- FJ882963.1 SARS coronavirus P2
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatcacaaaaaaaa--------------------------- AY304495.1 SARS coronavirus GZ50
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaa------------------ AY278488.2 SARS coronavirus BJ01
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaa-------------------- AY485278.1 SARS coronavirus Sino3-11
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaa-------------------- AY278491.2 SARS coronavirus HKU-39849
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaa AY357076.1 SARS coronavirus PUMC03
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaaaaa-------- AY357075.1 SARS coronavirus PUMC02
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY395002.1 SARS coronavirus LC5
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY395001.1 SARS coronavirus LC4
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY395000.1 SARS coronavirus LC3
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY394999.1 SARS coronavirus LC2
---------------------------------------------------------------------------------------------------- AY286320.4 SARS coronavirus ZJ01, partial genome
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gaga---------------------------------------- DQ182595.1 SARS coronavirus ZJ0301 from China
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgaca---------------------------------- AY559092.1 SARS coronavirus SinP5
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgaca---------------------------------- AY559088.1 SARS coronavirus SinP1
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttctagg-aagaatga------------------------------------ AY559091.1 SARS coronavirus SinP4
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaa--------------------------------- AY502924.1 SARS coronavirus TW11
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaa------------------ AY485277.1 SARS coronavirus Sino1-11
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaa---------------- AY654624.1 SARS coronavirus TJF
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaa-------------------------------- AY394850.2 SARS coronavirus WHU
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY394978.1 SARS coronavirus GZ-B
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaa-------------- AY279354.2 SARS coronavirus BJ04
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaa-------------- AY508724.1 SARS coronavirus NS-1
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaa----------------- AY278487.3 SARS coronavirus BJ02
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaa------------------ AY278490.3 SARS coronavirus BJ03
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY394995.1 SARS coronavirus HSZ-Cc
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY394994.1 SARS coronavirus HSZ-Bc
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY394985.1 SARS coronavirus HSZ-Bb
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY394986.1 SARS coronavirus HSZ-Cb
gtgt-aaaattaattttagtagtgctatccccatgtgatttta----cacattag-ggctcttccatatagg---------------------------- AY461660.1 SARS coronavirus SoD
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgaca---------------------------------- AY559094.1 SARS coronavirus Sin846
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaa------------------------------ AY595412.1 SARS coronavirus LLJ-2004
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgaca---------------------------------- AY559082.1 SARS coronavirus Sin852
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaa------------------- AY313906.1 SARS coronavirus GD69
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514423.1 SARS coronavirus wtic-MB strain SARS/VeroE6_lab/USA/WTic_c3P20/2009
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514422.1 SARS coronavirus wtic-MB strain SARS/VeroE6_lab/USA/WTic_c1.3P20/2010
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514421.1 SARS coronavirus wtic-MB strain SARS/VeroE6_lab/USA/WTic_c1.2P20/2010
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514419.1 SARS coronavirus wtic-MB strain SARS/VeroE6_lab/USA/WTic_c1P10/2009
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514418.1 SARS coronavirus wtic-MB strain SARS/VeroE6_lab/USA/WTic_c3P1/2009
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514415.1 SARS coronavirus wtic-MB strain SARS/VeroE6_lab/USA/WTic_c1.7P20/2010
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514413.1 SARS coronavirus wtic-MB strain SARS/VeroE6_lab/USA/WTic_c1.6P20/2010
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514409.1 SARS coronavirus wtic-MB strain SARS/VeroE6_lab/USA/WTic_c2P20/2009
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514408.1 SARS coronavirus wtic-MB strain SARS/VeroE6_lab/USA/WTic_c2P1/2009
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514404.1 SARS coronavirus wtic-MB strain SARS/VeroE6_lab/USA/WTic_c1.9P20/2010
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514400.1 SARS coronavirus wtic-MB strain SARS/VeroE6_lab/USA/WTic_c1.8P20/2010
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514399.1 SARS coronavirus wtic-MB strain SARS/VeroE6_lab/USA/WTic_c1.1P20/2010
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514398.1 SARS coronavirus wtic-MB strain SARS/VeroE6_lab/USA/WTic_c1.10P20/2010
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514397.1 SARS coronavirus wtic-MB strain SARS/VeroE6_lab/USA/WTic_c2P10/2009
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514396.1 SARS coronavirus wtic-MB strain SARS/VeroE6_lab/USA/WTic_c3P10/2009
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514394.1 SARS coronavirus wtic-MB strain SARS/VeroE6_lab/USA/WTic_c1P20/2009
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514392.1 SARS coronavirus wtic-MB strain SARS/VeroE6_lab/USA/WTic_c1.4P20/2010
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514388.1 SARS coronavirus wtic-MB strain SARS/VeroE6_lab/USA/WTic_c1.5P20/2010
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- FJ882938.1 SARS coronavirus wtic-MB
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- FJ882927.1 SARS coronavirus wtic-MB isolate P1pp1
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514420.1 SARS coronavirus ExoN1 strain SARS/VeroE6_lab/USA/ExoN1_c5P10/2009
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514417.1 SARS coronavirus ExoN1 strain SARS/VeroE6_lab/USA/ExoN1_c5.3P20/2010
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514416.1 SARS coronavirus ExoN1 strain SARS/VeroE6_lab/USA/ExoN1_c5.8P20/2010
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514414.1 SARS coronavirus ExoN1 strain SARS/VeroE6_lab/USA/ExoN1_c5P20/2009
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514412.1 SARS coronavirus ExoN1 strain SARS/VeroE6_lab/USA/ExoN1_c13P20/2009
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514411.1 SARS coronavirus ExoN1 strain SARS/VeroE6_lab/USA/ExoN1_c13P10/2009
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514410.1 SARS coronavirus ExoN1 strain SARS/VeroE6_lab/USA/ExoN1_c8P20/2009
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514406.1 SARS coronavirus ExoN1 strain SARS/VeroE6_lab/USA/ExoN1_c13P1/2009
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514405.1 SARS coronavirus ExoN1 strain SARS/VeroE6_lab/USA/ExoN1_c5.2P20/2010
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514403.1 SARS coronavirus ExoN1 strain SARS/VeroE6_lab/USA/ExoN1_c5.1P20/2010
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514402.1 SARS coronavirus ExoN1 strain SARS/VeroE6_lab/USA/ExoN1_c5.6P20/2010
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514401.1 SARS coronavirus ExoN1 strain SARS/VeroE6_lab/USA/ExoN1_c5.5P20/2010
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514395.1 SARS coronavirus ExoN1 strain SARS/VeroE6_lab/USA/ExoN1_c8P1/2009
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514393.1 SARS coronavirus ExoN1 strain SARS/VeroE6_lab/USA/ExoN1_c5.10P20/2010
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514391.1 SARS coronavirus ExoN1 strain SARS/VeroE6_lab/USA/ExoN1_c5.9P20/2010
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514390.1 SARS coronavirus ExoN1 strain SARS/VeroE6_lab/USA/ExoN1_c5.4P20/2010
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514389.1 SARS coronavirus ExoN1 strain SARS/VeroE6_lab/USA/ExoN1_c8P10/2009
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- FJ882930.1 SARS coronavirus ExoN1
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- FJ882926.1 SARS coronavirus ExoN1
gtgt-aaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- FJ882928.1 SARS coronavirus ExoN1 isolate P1pp1
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- JF292921.1 SARS coronavirus wtic-MB isolate c1P1
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- FJ882936.1 SARS coronavirus wtic-MB isolate P3pp2
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- FJ882932.1 SARS coronavirus wtic-MB isolate P3pp14
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- FJ882939.1 SARS coronavirus wtic-MB isolate P3pp16
gtgt-aaaattaattttagtagtg---------------------------------------------------------------------------- FJ882934.1 SARS coronavirus wtic-MB isolate P3pp29
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- FJ882947.1 SARS coronavirus wtic-MB isolate P3pp7
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- FJ882949.1 SARS coronavirus wtic-MB isolate P3pp23
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- FJ882946.1 SARS coronavirus wtic-MB isolate P3pp13
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- FJ882935.1 SARS coronavirus wtic-MB isolate P3pp21
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- FJ882937.1 SARS coronavirus wtic-MB isolate P3pp18
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- FJ882933.1 SARS coronavirus wtic-MB isolate P3pp6
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- JF292920.1 SARS coronavirus MA15 isolate d3om5
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- JF292919.1 SARS coronavirus MA15 isolate d3om4
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- JF292918.1 SARS coronavirus MA15 isolate d3om3
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- JF292917.1 SARS coronavirus MA15 isolate d3om2
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- JF292916.1 SARS coronavirus MA15 isolate d3om1
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- JF292912.1 SARS coronavirus MA15 isolate d4ym2
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- JF292911.1 SARS coronavirus MA15 isolate d4ym1
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- JF292908.1 SARS coronavirus MA15 isolate d2ym3
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- JF292907.1 SARS coronavirus MA15 isolate d2ym2
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- HQ890546.1 SARS coronavirus MA15 isolate d2om5
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- HQ890545.1 SARS coronavirus MA15 isolate d2om4
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- HQ890544.1 SARS coronavirus MA15 isolate d2om3
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- HQ890543.1 SARS coronavirus MA15 isolate d2om2
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- HQ890542.1 SARS coronavirus MA15 isolate d2om1
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- HQ890541.1 SARS coronavirus MA15 isolate d2ym1
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- JF292915.1 SARS coronavirus MA15 isolate d4ym5
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- JF292914.1 SARS coronavirus MA15 isolate d4ym4
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- JF292913.1 SARS coronavirus MA15 isolate d4ym3
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- JF292910.1 SARS coronavirus MA15 isolate d2ym5
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- JF292909.1 SARS coronavirus MA15 isolate d2ym4
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- JF292906.1 SARS coronavirus MA15 ExoN1 isolate d3om5
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- HQ890535.1 SARS coronavirus MA15 ExoN1 isolate d2om2
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- JF292905.1 SARS coronavirus MA15 ExoN1 isolate d3om4
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- HQ890540.1 SARS coronavirus MA15 ExoN1 isolate d3om2
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- HQ890539.1 SARS coronavirus MA15 ExoN1 isolate d3om1
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- HQ890538.1 SARS coronavirus MA15 ExoN1 isolate d2om5
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- JF292902.1 SARS coronavirus MA15 ExoN1 isolate d4ym4
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- HQ890537.1 SARS coronavirus MA15 ExoN1 isolate d2om4
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- HQ890533.1 SARS coronavirus MA15 ExoN1 isolate d4ym3
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- HQ890530.1 SARS coronavirus MA15 ExoN1 isolate d2ym5
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- HQ890528.1 SARS coronavirus MA15 ExoN1 isolate d2ym3
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- HQ890527.1 SARS coronavirus MA15 ExoN1 isolate d2ym2
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- HQ890526.1 SARS coronavirus MA15 ExoN1 isolate d2ym1
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- HQ890536.1 SARS coronavirus MA15 ExoN1 isolate d2om3
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- HQ890534.1 SARS coronavirus MA15 ExoN1 isolate d2om1
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- HQ890529.1 SARS coronavirus MA15 ExoN1 isolate d2ym4
gtgt-aaaattaattttagtag------------------------------------------------------------------------------ JF292904.1 SARS coronavirus MA15 ExoN1 isolate d3om3
gtgt-aaaattaattttagtag------------------------------------------------------------------------------ HQ890531.1 SARS coronavirus MA15 ExoN1 isolate d4ym1
gtgt-aaaattaattttagtag------------------------------------------------------------------------------ JF292903.1 SARS coronavirus MA15 ExoN1 isolate d4ym5
gtgt-aaaattaattttagtag------------------------------------------------------------------------------ HQ890532.1 SARS coronavirus MA15 ExoN1 isolate d4ym2
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- FJ882961.1 SARS coronavirus MA15 isolate P3pp5
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- FJ882958.1 SARS coronavirus MA15 isolate P3pp7
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- FJ882952.1 SARS coronavirus MA15 isolate P3pp4
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- FJ882945.1 SARS coronavirus MA15 isolate P3pp6
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- FJ882948.1 SARS coronavirus MA15 isolate P3pp3
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- FJ882957.1 SARS coronavirus MA15
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- FJ882943.1 SARS coronavirus MA15 ExoN1
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- FJ882953.1 SARS coronavirus MA15 ExoN1 isolate P3pp4
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- FJ882960.1 SARS coronavirus ExoN1 isolate P3pp34
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- FJ882955.1 SARS coronavirus ExoN1 isolate P3pp19
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- FJ882950.1 SARS coronavirus ExoN1 isolate P3pp60
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- FJ882940.1 SARS coronavirus ExoN1 isolate P3pp37
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- FJ882931.1 SARS coronavirus ExoN1 isolate P3pp12
gtgt-aaaattaattttagta------------------------------------------------------------------------------- FJ882956.1 SARS coronavirus ExoN1 isolate P3pp53
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- FJ882944.1 SARS coronavirus ExoN1 isolate P3pp23
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- FJ882954.1 SARS coronavirus ExoN1 isolate P3pp46
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- FJ882929.1 SARS coronavirus ExoN1 isolate P3pp1
gtgt-aaaattaattttagtagt----------------------------------------------------------------------------- FJ882941.1 SARS coronavirus ExoN1 isolate P3pp8
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- FJ882962.1 SARS coronavirus MA15 ExoN1 isolate P3pp10
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- FJ882951.1 SARS coronavirus MA15 ExoN1 isolate P3pp3
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- FJ882942.1 SARS coronavirus MA15 ExoN1 isolate P3pp5
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- FJ882959.1 SARS coronavirus MA15 ExoN1 isolate P3pp6
gtgtaaaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- KF514407.1 SARS coronavirus ExoN1 strain SARS/VeroE6_lab/USA/ExoN1_c5.7P20/2010
gtgtaaaaattaattttagtagt----------------------------------------------------------------------------- JF292922.1 SARS coronavirus ExoN1 isolate c5P1
gtgtaaaaattaattttagtagtgctatccccatgtgattttaa-------------------------------------------------------- JX162087.1 SARS coronavirus ExoN1 isolate c5P10
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- MK062182.1 SARS coronavirus Urbani isolate icSARS-C3-MA
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- MK062181.1 SARS coronavirus Urbani isolate icSARS-C3
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaa------------------------------- EU371564.1 SARS coronavirus BJ182-12
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaa------------------------------- EU371563.1 SARS coronavirus BJ182-8
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaa------------------------------- EU371561.1 SARS coronavirus BJ182b
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaa------------------------------- EU371560.1 SARS coronavirus BJ182a
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaa------------------------------- EU371562.1 SARS coronavirus BJ182-4
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaa------------------------------ AY559097.1 SARS coronavirus Sin3408L
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaa---------------------- DQ640652.1 SARS coronavirus GDH-BJH01
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY395003.1 SARS coronavirus ZS-C
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY394997.1 SARS coronavirus ZS-A
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY394996.1 SARS coronavirus ZS-B
gtgt-aaa-------------------------------------------------------------------------------------------- AY463060.1 SARS coronavirus ShanghaiQXC2
gtgt-aaa-------------------------------------------------------------------------------------------- AY463059.1 SARS coronavirus ShanghaiQXC1
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY394979.1 SARS coronavirus GZ-C
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttctaggagaaaatgaca---------------------------------- AY559090.1 SARS coronavirus SinP3
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgaca---------------------------------- AY772062.1 SARS coronavirus WH20
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatcac----------------------------------- FJ959407.1 SARS coronavirus isolate A001
---------------------------------------------------------------------------------------------------- AY686864.1 SARS coronavirus B039
---------------------------------------------------------------------------------------------------- AY686863.1 SARS coronavirus A022
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttctta---------------------------------------------- AY545914.1 SARS coronavirus isolate HC/SZ/79/03
---------------------------------------------------------------------------------------------------- AY572035.1 SARS coronavirus civet010
---------------------------------------------------------------------------------------------------- AY572034.1 SARS coronavirus civet007
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatc------------------------------------- AY515512.1 SARS coronavirus HC/SZ/61/03
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatc------------------------------------- AY572038.1 SARS coronavirus civet020
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatcacaaaaaaaaaaaaaaaa------------------- AY545918.1 SARS coronavirus isolate HC/GZ/32/03
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatcac----------------------------------- AY545916.1 SARS coronavirus isolate HC/SZ/266/03
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttctta---------------------------------------------- AY545915.1 SARS coronavirus isolate HC/SZ/DM1/03
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaa------------------------------ AY545919.1 SARS coronavirus isolate CFB/SZ/94/03
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatcacaaaaaaaaaaaaaaaaaaaaa-------------- AY545917.1 SARS coronavirus isolate HC/GZ/81/03
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY613950.1 SARS coronavirus PC4-227
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY613949.1 SARS coronavirus PC4-136
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY613948.1 SARS coronavirus PC4-13
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY613947.1 SARS coronavirus GZ0402
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaaaaaaaaaaaaaaaaaaaa----------- AY568539.1 SARS coronavirus GZ0401
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaat-------------------------------------- AY297028.1 SARS coronavirus ZJ01
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaaaaaa----------------------------- AY559083.1 SARS coronavirus Sin3408
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- MK062184.1 SARS coronavirus Urbani isolate icSARS-C7-MA
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgac----------------------------------- MK062183.1 SARS coronavirus Urbani isolate icSARS-C7
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaa--------------------------------- AY559089.1 SARS coronavirus SinP2
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag--------------------------------------------- AB257344.1 SARS coronavirus Frankfurt 1 genomic RNA, nearly complete genome, clone: persistent virus #21
gtgt-aaaattaattttagtagtgctatccccatgtgattttaatagcttcttag-gagaatgacaa--------------------------------- AY351680.1 SARS coronavirus ZMY 1
The number of A bases at the end of the sequence was
most often zero, next most often 2, and third most often 24, and the
maximum length of the poly(A) tail was 35 bases:
$ seqkit seq -s sars1.aln|tr -d -|rev|sed 's/[^a].*//'|awk '{print length}'|sort|uniq -c|sort -nk2
117 0
17 1
58 2
3 3
7 4
4 5
1 6
1 8
1 11
2 13
2 15
2 16
4 17
1 18
1 19
3 21
38 24
1 27
1 35
In a paper titled "The Architecture of SARS-CoV-2 Transcriptome", the authors wrote: "We recently developed software to measure the length of poly(A) tail from DRS data (Y. Choi and H.C., unpublished data). Using this software, we confirm that, like other CoVs, SARS-CoV-2 RNAs carry poly(A) tails (Figures 4A-4B). The tail of viral RNAs is 47 nt in median length. The full-length gRNA has a relatively longer tail than sgRNAs. Notably, sgRNAs have two tail populations: a minor peak at ∼30 nt and a major peak at ∼45 nt (Figure 4B, arrowheads). Wu et al., 2013 previously observed that the poly(A) tail length of bovine CoV mRNAs changes during infection: from ∼45 nt immediately after virus entry to ∼65 nt at 6-9 hours post-infection and ∼30 nt at 120-144 hours post-infection. Thus, the short tails of ∼30 nt observed in this study may represent aged RNAs that are prone to decay." [https://www.cell.com/cell/fulltext/S0092-8674%2820%2930406-2):]
On GISAID, the SARS2 sequence with the oldest collection date is EPI_ISL_402123 (IPBCAMS-WH-01), which is listed as being collected on 2019-12-24. The sample was described in a paper by Ren et al. which was only published in May 2020. [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7147275/] However there's also a blog post about the procedure of sequencing the sample at which was posted at WeChat on January 30th[ https://freewechat.com/a/MzAxMjMyMDk0Ng%3d%3d/2650112053/1]
Compared to the third version of Wuhan-Hu-1 at GenBank, IPBCAMS-WH-01 has three nucleotide changes within the ORF1a gene and it's missing the last four bases of the poly(A) tail:
$ curl https://download.cncb.ac.cn/gwh/Viruses/Severe_acute_respiratory_syndrome_coronavirus_2_IPBCAMS-WH-01_GWHABKF00000000/GWHABKF00000000.genome.fasta.gz|gzip -dc|cat - <(curl 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=MN908947.3')|mafft --quiet ->temp.fa
$ Rscript -e 'm=t(as.matrix(Biostrings::readDNAStringSet("temp.fa")));pick=which(rowSums(m!=m[,1])>0);write.table(cbind(m[pick,],pick),quote=F,row.names=F,col.names=F)'
G A 3778
G A 8388
A T 8987
- A 29900
- A 29901
- A 29902
- A 29903
However the three nucleotide changes are probably just some kind of artifacts of sequencing or assembly. In a paper about early human cases, babarlelephant wrote: "The WHO report reanalyzed several of the early sequences and found that the three mutations of IPBCAMS-WH-01 were not supported by the raw reads (which are not publicly available)." [https://zenodo.org/record/6715352]
In one paper where there's a list of mutations in 66 early SARS2 sequences, IPBCAMS-WH-01 was the only sequence with any mutation out of A3778G, A8388G, and T8987A. [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7834520/] I found a text file which includes a list of mutations in about 80,000 GISAID sequences until September 2020. [https://preearth.net/pdfs/ids-cdate-sdate-name-country-mutations-77827.txt] Apart from IPBCAMS-WH-01, there were only 4 other sequences with A3778G, 2 with A8388G, and 4 with T8987A, but all of them had a collection date in March 2020 or later:
$ wget -q https://preearth.net/pdfs/ids-cdate-sdate-name-country-mutations-77827.txt
$ for x in A3778G A8388G T8987A;do awk '{$1=$1}1' FS='\n *' RS= OFS=\; ids-cdate-sdate-name-country-mutations-77827.txt|grep -v EPI_ISL_402123|grep $x|wc -l|sed s/^/$x\ /;done
A3778G 4
A8388G 2
T8987A 4
The paper by Ren et al. said: "Quality control processes included removal of low-complexity reads by bbduk (entropy = 0.7, entropy-window = 50, entropy k = 5; version: January 25, 2018),[11] adapter trimming, low-quality reads removal, short reads removal by Trimmomatic (adapter: TruSeq3-SE.fa:2:30:6, LEADING: 3, TRAILING: 3, SLIDING WINDOW: 4:10, MINLEN: 70, version: 0.36),[12] host removal by bmtagger (using human genome GRCh38 and yh-specific sequences as reference),[13] and ribosomal reads removal by SortMeRNA (version: 2.1b).[14]" Wu et al. didn't describe removing low-complexity reads, host reads, or ribosomal reads, but they just masked human reads with N bases in the reads they uploaded to the SRA.
The 5' UTR is also identical to Wuhan-Hu-1 in IPBCAMS-WH-03, 04, and 05, but IPBCAMS-WH-02 has 6 nucleotide changes in the region 104-124 (so it might be some kind of an error):
$ curl 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=MN908947,MT019529,MT019530,MT019531,MT019532,MT019533'|mafft --quiet ->ren.fa
$ aldi2()(cat "$@">/tmp/aldi;Rscript --no-init-file -e 'm=t(as.matrix(Biostrings::readAAStringSet("/tmp/aldi")));diff=which(rowSums(m!=m[,1])>0);split(diff,cumsum(diff(c(-Inf,diff))!=1))|>sapply(\(x)c(if(length(x)==1)x else paste0(x[1],"-",tail(x,1)),apply(m[x,,drop=F],2,paste,collapse="")))|>t()|>write.table(quote=F,sep=";",row.names=F)')
$ R -e 'if(!require("BiocManager"))install.packages("BiocManager");BiocManager::install("Biostrings")'
[...]
$ for i in {2..6};do (seqkit head -n1 ren.fa;seqkit range -r$i:$i ren.fa)|sed 's/,.*//;s,/20.*,,;s,.*\(/\| \),>,'|aldi2;echo;done
;Wuhan-Hu-1;IPBCAMS-WH-01
3778;a;g
8388;a;g
8987;t;a
29900-29903;aaaa;----
;Wuhan-Hu-1;IPBCAMS-WH-02
104;t;a
111-112;tt;cg
119-120;ct;gc
124;g;a
29890-29903;aaaaaaaaaaaaaa;--------------
;Wuhan-Hu-1;IPBCAMS-WH-03
6996;t;c
29900-29903;aaaa;----
;Wuhan-Hu-1;IPBCAMS-WH-04
29891-29903;aaaaaaaaaaaaa;-------------
;Wuhan-Hu-1;IPBCAMS-WH-05
7866;g;t
29884-29903;aaaaaaaaaaaaaaaaaaaa;--------------------
If you search the NCBI's sequence read archive for something like
sars nanopore, almost all results employ a PCR-based
sequencing protocol where RNA is converted to cDNA and then segments of
cDNA are amplified with PCR. The amplicon length is around 400 bases in
the ARCTIC protocol and around 1,200 bases in the MIDNIGHT protocol.
However in a variant of nanopore sequencing called direct RNA nanopore sequencing, the conversion to cDNA is not needed, and the read lengths can exceed 10,000 bases. A paper from 2019 described using direct RNA nanopore sequencing to sequence almost the entire length of a coronavirus in a single piece. [https://genome.cshlp.org/content/29/9/1545.full]
When I searched the SRA for direct rna nanopore sars,
there were only 130 results.
[https://www.ncbi.nlm.nih.gov/sra/?term=direct+rna+nanopore+sars]
In many runs, the maximum read length was only a few thousand bases, but
I found one run where the longest SARS2 read was over 10,000 bases long.
[https://www.ncbi.nlm.nih.gov/sra/?term=SRR11300652]
The longest read in the entire run was about 52,000 bases, but the
longest read that aligned against SARS2 was 14,191 bases long and it had
about 93% identity with Wuhan-Hu-1:
$ curl -s 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=MN908947.3'>sars2.fa $ wget ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR113/052/SRR11300652/SRR11300652_1.fastq.gz [...] $ brew install blast seqkit [...] $ makeblastdb -in sars2.fa -dbtype nucl [...] $ seqkit fq2fa SRR11300652_1.fastq.gz|seqkit sort -lr|seqkit head -n20|blastn -db sars2.fa -outfmt '6 qseqid pident length qlen qstart qend sstart send'|tr \\t ' ' SRR11300652.328671 92.657 14191 13831 1 13753 16008 29873
WIV02 to WIV07 are samples of early COVID patients that were published in the same paper as RaTG13. They're among the relatively rare sequences of SARS-CoV-2 at GenBank that were de-novo assembled. But only WIV04 matches the full 5' end and only WIV04 includes a poly(A) tail:
$ curl 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id='MN{908947,9965{27..31}}>wiv.fa
$ seqkit replace -p'.* (.*?),.*' -r\$1 wiv.fa|seqkit subseq -r1:200|mafft --quiet -|seqkit subseq -r1:60|seqkit fx2tab|column -t
Wuhan-Hu-1 attaaaggtttataccttcccaggtaacaaaccaaccaactttcgatctcttgtagatct
WIV02 ---------------------------------aaccaactttcgatctcttgtagatct
WIV04 attaaaggtttataccttcccaggtaacaaaccaaccaactttcgatctcttgtagatct
WIV05 ------------taccttcccaggtaacaaaccaaccaactttcgatctcttgtagatct
WIV06 --------------ccttcccaggtaacaaaccaaccaactttcgatctcttgtagatct
WIV07 -------------accttcccaggtaacaaaccaaccaactttcgatctcttgtagatct
$ seqkit replace -p'.* (.*?),.*' -r\$1 wiv.fa|seqkit subseq -r-200:-1|mafft --quiet -|seqkit subseq -r -60:-1|seqkit fx2tab|column -t
Wuhan-Hu-1 attttaatagcttcttaggagaatgacaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaaa
WIV02 attttaatagcttct---------------------------------------------
WIV04 attttaatagcttcttaggagaatgacaaaaaaaaaaaaaaaaaaaaa------------
WIV05 attttaatagcttcttaggag---------------------------------------
WIV06 attttaatagcttcttaggagaatg-----------------------------------
WIV07 attttaatagcttcttaggagaatgac---------------------------------
There were 230 results at GenBank when I searched for
txid2697049 complete genome megahit (where the taxid is the
ID of SARS-CoV-2).
[https://www.ncbi.nlm.nih.gov/nuccore/?term=megahit+txid2697049+25000%3A35000%5Bsequence+length%5D]
In all results I checked, the comment field mentioned that the sequence
was assembled with MEGAHIT.
However only 36 of the results matched the full 5' end of Wuhan-Hu-1 so that they didn't have extra bases before it. 3 results had extra bases before the start of Wuhan-Hu-1:
$ esearch -db nuccore -query 'txid2697049 complete genome megahit'|efetch -format fasta>mega.fa
$ seqkit subseq -r1:200 mega.fa|mafft --quiet -|seqkit seq -s|cut -c-40|sort|uniq -c|sort -r
194 ----------------------------------------
36 ----------------------attaaaggtttatacctt
8 ------------------------------------cctt
6 ---------------------------------------t
5 ----------------------------ggtttatacctt
5 -----------------------------gtttatacctt
5 ---------------------------------atacctt
4 -----------------------ttaaaggtttatacctt
3 ------------------------taaaggtttatacctt
3 --------------------------aaggtttatacctt
3 -------------------------------------ctt
2 ------------------------------tttatacctt
2 ----------------------------------tacctt
2 -----------------------------------acctt
1 tgtgctcttccgatctggagggattaaaggtttatacctt
1 ---------------------ggttttaggttyatacctt
1 ---------------------gattaaaggtttatacctt
1 ----------------------cttaaaggtttatacctt
1 ----------------------atcaaaggtttatacctt
1 -------------------------aaaggtttatacctt
1 --------------------------------------tt
156 out of 230 sequences were missing the poly(A) tail. And 10 sequences had a poly(A) tail longer than 33 bases:
$ seqkit subseq -r-200:-1 mega.fa|mafft --quiet -|seqkit subseq -r-70:-1|seqkit seq -s|sed 's/-.*$//'|awk '{a[length]++}END{for(i in a)print i,a[i]}'
0 156
1 5
2 1
3 1
5 4
6 1
7 1
8 1
9 1
10 1
11 3
12 6
13 1
14 1
16 1
17 6
18 5
19 1
20 3
21 5
26 1
27 1
29 2
32 1
33 11
69 1
70 9
In 2022, someone who called themselves "a mathematician from Hamburg, who would like to remain unknown" published a paper titled "Structural analysis of sequence data in virology: An elementary approach using SARS-CoV-2 as an example". [https://impfen-nein-danke.de/u/structural_analysis_of_sequence_data_in_virology.pdf, https://impfen-nein-danke.de/u/structural_analysis_of_sequence_data_in_virology_tables_and_figures.pdf]
When the mathematician from Hamburg aligned Wu et al.'s reads with
BBMap against reference genomes of HIV-1, hepatitis delta, and measles,
he was able to get a large number of aligned reads, but that's because
he ran BBMap with the minratio=0.1 parameter which caused
it to tolerate a very high number of mismatches, so the most common
length of his aligned reads was about 35-40 bases, and the average error
rate among the reads was about 40%. And when he filtered his reads with
reformat.sh, he also used a low required minimum length
(37) and an extremely low required identity percent (60%). His paper
says that he ran BBMap with "relatively unspecific
settings", but actually he ran it with extremely liberal
alignment criteria:
Basically, we mapped the paired-end reads (2x151 bp) with BBMap [23] to the reference sequences we considered (Tables and Figures: Table 3) using relatively unspecific settings. We then varied the minimum length (M1) and minimum (nucleotide) identity (M2) with reformat.sh to obtain corresponding subsets of the previously mapped sequences with appropriate quality. Increasing minimum length M1 or minimum nucleotide identity M2 thereby increases the significance of the respective mapping. Subsequently, we formed consensus sequences with the respective subsets of selected quality with respect to the selected reference. We set all bases with a quality lower than 20 to "N" (unknown). A quality of 20 means an error rate of 1% per nucleotide, which can be considered sufficient in the context of our analyses. Finally, the assessment of the agreement between reference and consensus sequences was performed using BWA [24], Samtools [25], and Tablet [26]. The ordered pair (M1; M2) = (37; 0.6) was just chosen to give error rates F1 and F2, respectively, of less than 10% for reference LC312715.1. The results of all calculations performed are shown in Tables and Figures: Table 4. The calculations show the highest significance for the choice of the ordered pair (37; 0.6), which can be seen from the highest error rates in each case. Comparable significance is provided by the ordered pairs (47; 0.50) and (25; 0.62). While the genome sequences associated with coronaviruses show error rates approximately above 10% for all ordered pairs considered (M1; M2), the error rates of the two sequences LC312715.1 (HIV) and NC_001653.2 (Hepatitis delta) are below 10% and decrease further for the ordered pairs (32; 0.60) and (30; 0.60).
From the figure below, you can see that the mode length was around 150 bases for the reads which aligned against Wuhan-Hu-1 (MN908947.3), but the mode length was around 35-40 bases for the reads which aligned against HIV-1, hepatitis delta, measles, and randomly generated sequences:

I don't understand why the figure above shows that there's some reads
shorter than 37 bases even though the caption indicates that reads
shorter than 37 bases were filtered out with
reformat.sh.
And I also don't understand why the figure above shows that there
were reads longer than 151 bases which aligned against Wuhan-Hu-1,
because Wu et al.'s raw reads don't include any reads longer than 151
bases. Maybe the mathematician from Hamburg counted gaps in the middle
of the aligned reads as part of the reads, or maybe he calculated read
lengths based on the CIGAR strings so that he added together all numbers
in the CIGAR string without skipping deletions relative to the
reference. His paper indicated that he ran BBMap with the
maxindel=0 parameter which doesn't allow indels, but it
might be that he forgot to include the parameter in the run where he
used Wuhan-Hu-1 as the reference, since there doesn't seem to be reads
longer than 151 bases in the plots for other reference sequences
above.
In the plot for SARS-CoV-2 above, there's two peaks for the length of
the aligned reads, where the first peak around length 37 looks almost as
high as the second peak around length 150, but it's misleading that the
y-axis is cut off so you can't see the full height of the second peak. I
tried aligning Wu et al.'s raw reads against Wuhan-Hu-1 so that I ran
mapPacBio.sh and reformat.sh with the same
parameters as the mathematician from Hamburg, but I got about 11 times
as many aligned reads of length 151 as reads of length 37:

So you can compare my plot above to the plot by the mathematician by Hamburg to see how misleading it was that he cut off the y-axis, especially since it's the long reads with length around 151 that actually matched the virus reference sequences, and the short reads with length around 30-50 bases consisted of spurious matches that would have remained unaligned if BBMap was ran with regular parameters.
Next I tried aligning Wu et al.'s reads against other reference
genomes, where I again ran BBMap and reformat.sh with the
same parameters as the mathematician from Hamburg:
brew install --cask miniconda;conda init ${SHELL##*/}
conda install -c bioconda bbmap
brew install fastp
curl -s 'https://www.ebi.ac.uk/ena/portal/api/filereport?accession=SRR10971381&result=read_run&fields=fastq_ftp&format=tsv&download=true&limit=0'|sed 1d|cut -f2|tr \; \\n|sed s,^,ftp:,,|xargs wget -q
fastp -i SRR10971381_1.fastq.gz -I SRR10971381_2.fastq.gz -o SRR10971381_1.trim.fq.gz -O SRR10971381_2.trim.fq.gz # trim adapter sequences, trim low-quality ends of reads, discard reads shorter than 15 bases, discard reads with 5 or more `N` letters
printf %s\\n MN908947.3\ sars2 LC312715.1\ hiv NC_001653.2\ hepatitis AF266291.1\ measles>refs
cat refs|while read l m;do curl "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=$l">ref.$m.fa;done
gshuf -rn30000<<<$'A\nC\nG\nT'|tr -d \\n|awk 'BEGIN{print">random"}1'>ref.random.fa
for x in $(cut -d\ -f2 refs) random;do mapPacBio.sh ref=ref.$x.fa in=SRR10971381_1.trim.fq.gz in2=SRR10971381_2.trim.fq.gz outm=$x.mapped.sam vslow k=8 maxindel=0 minratio=0.1;done
for x in $(cut -d\ -f2 refs) random;do reformat.sh in=$x.mapped.sam out=$x.filtered.sam minlength=37 idfilter=60 ow=t;done
for x in $(cut -d\ -f2 refs) random;do awk -F\\t '/NM:i:/{l=length($10);sub(/.*NM:i:/,"");sub("\t.*","");a[l]+=$0;n[l]++}END{for(i in a)print i,n[i],100*a[i]/n[i]/i}' OFMT=%.1f $x.filtered.sam|sed s/^/$x\ /;done>lengthcounts
The plot below shows the result of my BBMap alignment. If you compare SARS2 to HIV-1, hepatitis delta, or the random sequence, they all have a similar mismatch rate and similar number of aligned reads at the lowest read lengths, but after around length 55, the mismatch rate of SARS2 starts to drop until it reaches about 2% at length 151. However the mismatch rate of the other sequences remains at around 40% even for longer reads:

Hepatitis delta has an extremely short genome of only about 1,700 bases, so in the plot above, hepatitis delta gets less aligned reads at lengths 37 to 60 than the longer genomes. I don't know why the difference between hepatitis delta and the longer genomes is not bigger though, even though SARS2 and the random 30-kb genome are almost 20 times longer than hepatitis delta, but it might be because the longer sequences are more likely to include multimapped reads which I omitted from this analysis. Or it might be because I ran BBMap with such loose alignment criteria that even for a short reference genome, random short reads are likely to get aligned against some part of the genome.
(The number of mismatches is indicated by the NM tag in
the SAM file format. For some reason BBMap doesn't include the
NM tag for multimapped reads, which are reads that can map
to two or more spots in the reference genome and which are indicated by
a zero in the fifth column of the SAM file. Therefore I omitted
multimapped reads from the plot above in order to calculate the average
number of mismatches correctly.)
Here's R code for generating the plot above:
library(ggplot2)
t=read.table("lengthcounts")
t2=rbind(t,expand.grid(V1=unique(t$V1),V2=unique(t$V2),V3=0,V4=NA))
t=t2[!duplicated(t2[,1:2]),]
group=factor(t$V1,levels=unique(t$V1))
color=c("black",hcl(c(0,210,120)+15,60,55),"gray60")
ybreak=1e4
ybreak2=8
xstart=xbreak*floor(min(t$V2)/xbreak)
xend=xbreak*ceiling(max(t$V2)/xbreak)
yend2=48
zeroshift=1
log=log10(t$V3)+zeroshift
log[log==-Inf]=0
yend=ceiling(max(log))
ylabel=c(0,1,10,100,paste0("1e",3:ceiling(max(log-zeroshift))))
mismatch=t$V4*yend/yend2
label=c("SARS2 (MN908947.3) (30kb)","HIV-1 (LC312715.1) (9kb)","Hepatitis delta (NC_001653.2) (2kb)","Measles (AF266291.1) (16kb)","Random sequence (30kb)")
ystep=yend/15
labels=data.frame(x=.25*(xend-xstart),y=seq(.3*yend,by=-ystep,length.out=length(label)),label)
ggplot(t,aes(x=V2,y=log,color=group))+
geom_line(linewidth=.3)+
geom_point(aes(y=mismatch),size=.2)+
geom_label(data=labels,aes(x=x,y=y,label=label),fill=alpha("white",.7),label.r=unit(0,"lines"),label.padding=unit(.04,"lines"),label.size=0,color=color,size=2.2,hjust=0)+
scale_x_continuous(limits=c(xstart,xend),breaks=seq(0,xend,xbreak),expand=c(0,0))+
scale_y_continuous(limits=c(0,yend),breaks=c(0,zeroshift+seq(0,5,1)),labels=ylabel,expand=expansion(mult=c(.02,0)),sec.axis=sec_axis(trans=~.*yend2/yend,breaks=seq(0,yend2,ybreak2),name="Percentage of mismatches (dots)"))+
labs(x="Length of read",y="Number of aligned reads (lines)")+
scale_color_manual(values=color)+
theme(
axis.text=element_text(size=6,color="black"),
axis.ticks=element_blank(),
axis.ticks.length=unit(0,"in"),
axis.title=element_text(size=8),
legend.position="none",
panel.background=element_rect(fill="white"),
panel.border=element_rect(color="gray85",fill=NA,linewidth=.4),
panel.grid.major=element_line(color="gray85",linewidth=.2),
panel.grid.minor=element_blank(),
plot.background=element_rect(fill="white"),
plot.margin=margin(.05,.15,.05,.1,"in")
)
ggsave("1.png",width=4,height=2.5)
The code below selects reads that aligned against SARS2, that were at
least 37 bases long so they survived the filtering by
reformat.sh, and that are not multimapped reads so they are
not missing the NM tag. The average number of mismatches
per read was 37.5% for reads of length 37 and 2.0% for reads of length
151, and there were a total of 4,856 aligned reads for length 37 and
62,766 aligned reads for length 151:
$ awk -F\\t '/NM:i:/{l=length($10);sub(/.*NM:i:/,"");sub("\t.*","");a[l]+=$0;n[l]++}END{for(i in a)print i,n[i],100*a[i]/n[i]/i}' OFMT=%.1f sars2.filtered.sam|(gsed -u 4q;echo ...;tail -n4)
37 4856 37.5
38 5010 38.0
39 3896 37.8
40 4363 38.2
...
148 2316 2.3
149 6084 2.0
150 32431 2.2
151 62766 2.0
However in the case of the reads that got aligned against the HIV-1 reference genome, there were only 14 aligned reads of length 151 and even those reads had an average mismatch rate of about 41%:
$ awk -F\\t '/NM:i:/{l=length($10);sub(/.*NM:i:/,"");sub("\t.*","");a[l]+=$0;n[l]++}END{for(i in a)print i,n[i],100*a[i]/n[i]/i}' OFMT=%.1f hiv.filtered.sam|(gsed -u 4q;echo ...;tail -n4)
37 4460 37.2
38 4471 37.5
39 3584 37.4
40 3843 37.8
...
148 2 41.2
149 3 40.7
150 16 41.1
151 14 40.6
The page below from the paper by the mathematician from Hamburg is a bit confusing, because in the table at the top he uses the term "short reads" to refer to sequencing reads with a short length, but at the end of the paragraph I highlighted he uses the term "short reads" to refer to sequencing reads in general, even though the set of about 120,000 reads he referred to mostly consisted of long reads with length above 100 bases. So in the last paragraph below, what he meant is that the reads which actually matched SARS2 were mostly long reads:

The mathematician from Hamburg wrote that among the reads with a
maximum length of 100 bases, the average mismatch rate was about 30%,
but that's because he ran BBMap with the minratio=0.1
parameter which tolerated an extremely high number of mismatches.
However there's also some reads with a short length which genuinely
match SARS2, and he would've been able to differentiate them from the
spurious matches if he ran BBMap with the default parameters, or if he
used reformat.sh to filter out reads with a high mismatch
rate, or if he simply looked at which reads got aligned when he ran
Bowtie2 with the default settings:
$ curl 'https://www.ebi.ac.uk/ena/portal/api/filereport?accession=SRR10971381&result=read_run&fields=fastq_ftp'|sed 1d|cut -f2|tr \; \\n|sed s,^,ftp://,|xargs wget [...] $ fastp -i SRR10971381_1.fastq.gz -I SRR10971381_2.fastq.gz -o SRR10971381_1.fq.gz -O SRR10971381_2.fq.gz [...] $ curl -s 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=MN908947.3'>sars2.fa $ bow()([[ -e $1.1.bt2 ]]||bowtie2-build --threads 3 "$1" "$1";bowtie2 --no-unal -p3 -x "$@") $ bow sars2.fa -1 SRR10971381_1.fq.gz -2 SRR10971381_2.fq.gz|samtools sort -@3 ->wu.bam [...] $ samtools view wu.bam|ruby -ane'puts [$F[0],$F[3],$F[5],$F[9],$F.grep(/NM:i/)[0][/\d+/]]*" "'|awk 'length($4)<=100'|head -n3|column -t SRR10971381.21679314 10 68M1I6M TGTTACCTTCCCAGGTAACAAACCAACCAACTTTCGATCTCTTGTAGATCTGTTCTCTAAACGAACTTATGCACT 6 SRR10971381.21679314 10 68M1I6M GGGTACCTTCCCAGGTAACAAACCAACCAACTTTCGATCTCTTGTAGATCTGTTCTCTAAACGAACTTATGCACT 7 SRR10971381.11034571 175 89M TGAATCTTCTGCAGGCTGCTTACGGTTTCGTCCGTGTTGCAGCCGATCATCAGCACATCTAGGTTTCGTCCGGGTGTGACCGAAAGGTG 3
The last column above shows the number of mismatches in each read. When I ran Bowtie2 with the default settings and I filtered out reads longer than 100 bases, the average mismatch rate was only about 9%:
$ samtools view wu.bam|ruby -ane'puts [$F[0],$F[3],$F[5],$F[9],$F.grep(/NM:i/)[0][/\d+/]]*" "'|awk 'length($4)<=100{x+=$5}END{print 100*x/NR}'
9.0812
When I tried aligning Wu et al.'s reads with Bowtie2, I got zero
reads that aligned against HIV-1, hepatitis delta, measles, or the
random sequence. I got 121,779 reads which aligned against SARS2, which
was identical to the figure reported by the mathematician from Hamburg,
because both of us did trimming with fastp using the
default parameters.
With Bowtie2, the shortest reads that aligned against SARS2 were 31 bases long. There were less than a hundred reads at each length below 89, and their error rate was much lower than in the case of BBMap. However after around length 100-120, the results of Bowtie2 start to converge with BBMap:

I ran BBMap with the same parameters as the mathematician from
Hamburg (vslow k=8 maxindel=0 minratio=0.1), but if I
would've ran it with the default parameters instead, the results
would've been similar to Bowtie2.
I ran Bowtie2 with the default parameters:
bowtie2-build sars2.fa{,}
bowtie2 -p3 -x sars2.fa -1 SRR10971381_1.trim.fq.gz -2 SRR10971381_2.trim.fq.gz --no-unal|samtools sort -@2 ->sorted30.bam
I created a random 30,000-base reference sequence and a thousand random reads for each length between 30 and 150. When I ran BBMap with the same parameters as the mathematician from Hamburg, 372 out of 1000 reads of length 30 got aligned, but I got zero aligned reads for each length above 70. The proportion of mismatches was around 40% for most read lengths but 59% for length 70. This time I didn't get any multimapped reads so I didn't have to exclude them:
$ gshuf -e A -e C -e G -e T -rn30000|printf %s\\n \>ref $(paste -sd\\0 -)>ref.fa
$ n=1000;for i in {30..150};do gshuf -e A -e C -e G -e T -rn$[i*n]|awk '{ORS=NR%i==0?RS:""}1' i=$i|paste <(seq $n|sed s/^/$i./) -;done|seqkit tab2fx>reads.fa
$ bbmap.sh ref=ref.fa in=reads.fa outm=reads.mapped.sam vslow k=8 maxindel=0 minratio=0.1
[...]
$ awk -F\\t '/^[^@]/{l=length($10);sub(/.*NM:i:/,"");sub(/\t.*/,"");a[l]+=$0;n[l]++}END{for(i in a)print i,n[i],100*a[i]/n[i]/i}' OFMT=%.1f reads.mapped.sam
30 372 40.6
31 312 40.8
32 230 42.1
33 196 41.0
34 180 41.5
35 137 41.6
36 104 41.3
37 89 41.2
38 73 41.5
39 53 41.5
40 41 39.9
41 29 43.5
42 24 40.3
43 24 41.4
44 15 43.2
45 14 40.8
46 13 41.3
47 10 41.7
48 6 41.7
49 7 42.6
50 3 40
51 3 43.8
52 5 43.8
53 3 40.9
54 5 41.5
55 2 42.7
56 3 42.3
57 1 40.4
70 1 58.6
When I filtered out reads with indentity percent below 70, I was left with only 39 reads:
$ reformat.sh in=reads.mapped.sam out=reads.filtered.sam idfilter=70 ow=t
[...]
$ awk -F\\t '/^[^@]/{l=length($10);sub(/.*NM:i:/,"");sub(/\t.*/,"");a[l]+=$0;n[l]++}END{for(i in a)print i,n[i],100*a[i]/n[i]/i}' OFMT=%.1f reads.filtered.sam
30 17 28.6
31 11 28.4
32 2 23.4
33 2 27.3
34 5 28.2
35 1 28.6
38 1 28.9
When I ran BBMap with the default parameters instead of the parameters used by the mathematician from Hamburg, zero of the random reads mapped to the random reference genome (which is in fact what should happen when you're running an aligner with sensible parameters).
When I googled for "minratio=0.1" "bbmap", I found that
the BBTools user guide included the command
mapPacBio.sh in=reads.fq out=mapped.sam vslow k=8 maxindel=200 minratio=0.1,
with the comment that the command can be used to "map with super-high sensitivity (useful for
very-low-quality data, or remote homologies)".
[https://jgi.doe.gov/data-and-tools/software-tools/bbtools/bb-tools-user-guide/bbmap-guide/]
A list of shell commands used by the mathematician from Hamburg
featured this BBMap command:
mapPacBio.sh in=SRR10971381_1.fastq in2=SRR10971381_2.fastq outm=mapped.sam vslow k=8 maxindel=0 minratio=0.1.
So maybe he copied the command from the BBTools user guide, because he
even used the same output file name mapped.sam, he listed
the parameters in the same order, and he used mapPacBio.sh
instead of bbmap.sh.
The value for the K-mer length can range from 8 to 15, where the
default value is k=13 and lower values are more sensitive.
So the mathematician from Hamburg used the most sensitive possible value
for the K-mer length.
The minratio parameter is no longer listed in the help
message of BBMap, but I think it's because it has the same effect as the
minid parameter but it uses a different scale, so it's
basically redundant and the developer of BBMap recommended using
minid instead. The default value for minid is
0.76 which is approximately equal to minratio=0.56. The
developer of BBMap wrote the following:
[https://www.biostars.org/p/376782/]
Sorry for the confusion; they set the same thing, just with different units. "minratio" is the internal number using the internal scoring system; if you use the "minid" flag it converts it to the equivalent minratio. So if you set "minid=0.76" then it will be converted to approximately "minratio=0.56" and then the minratio variable will be set to 0.56. So there is no reason to set both of them. If you do set both of them, the last value will be used. I recommend minid over minratio because it is easier to read, but they accomplish the same thing.
"minid" is approximate since it adjusts minratio, or the ratio of the maximum possible internal score, but the internal score is not directly related to percent identity because of different penalties for substitutions, deletions, and insertions, which vary by length. If you need an exact %identity cutoff, use "idfilter=0.76" which will eliminate all alignments below 76% identity, as a postfilter. "minratio/minid" actually affect mapping, and prevent the creation of any alignments with an internal score below "minratio". As such, a high value of minid will make the program faster and a low value will make it slower. Whereas "idfilter" does not affect speed.
That said, I don't recommend using "idfilter" unless you have a good reason (like, a paper with a graph showing %identity cutoffs, for clarity; not for something like variant calling). "minid/minratio" are proportional to the internal score which should better model actual mutations or evolution, by trying to minimize the number of independant events rather than the edit distance; "idfilter=0.5", for example, will disallow a 100bp read from mapping with a 400bp deletion in the middle, since that would be only 20% identity. "minid=0.5" would allow this mapping because the internal score would actually still be quite high (maybe 0.9 of the maximum possible ratio).
The mathematician from Hamburg wrote:
Alignment with the nucleotide database on 05/12/2021 showed a high match (98.85%) with "Homo sapiens RNA, 45S pre-ribosomal N4 (RNA45SN4), ribosomal RNA" (GenBank: NR_146117.1, dated 04/07/2020). This observation contradicts the claim in [1] that ribosomal RNA depletion was performed and human sequence reads were filtered using the human reference genome (human release 32, GRCh38.p13). Of particular note here is the fact that the sequence NR_146117.1 was not published until after the publication of the SRR10971381 sequence library considered here.
But actually the date that is shown in the top right corner at the NCBI's website is the date when the entry was modified, and the original publication date of NR_146117 is in 2017. You can see the original publication date if you click the small "GenBank" menu in the top left corner and select "Revision History". [https://www.ncbi.nlm.nih.gov/nuccore/NR_146117.1?report=girevhist]
When I ran BBMap with the same extremely liberal criteria as the
mathematician from Hamburg, I got over 200,000 reads which aligned
against HIV but all of them had a high error rate. The lowest error rate
was 10% in a read that was 33 bases long, but the first 23 bases were
skipped by the aligner (23S), the next base had a mismatch
(1X), and only the last 9 bases matched the reference
(9=). Such a short match would be ignored with regular
alignment criteria:
$ brew install fastp
$ brew install --cask miniconda;conda init ${SHELL##*/};conda install -c bioconda bbmap
$ curl -s "https://www.ebi.ac.uk/ena/portal/api/filereport?accession=SRR10971381&result=read_run&fields=fastq_ftp"|sed 1d|cut -f2|tr \; \\n|sed s,^,ftp://,|xargs wget
$ fastp -i SRR10971381_1.fastq.gz -I SRR10971381_2.fastq.gz -o SRR10971381_1.fq.gz -O SRR10971381_2.fq.gz
$ curl -s 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=LC312715'>ref.hiv.fa
$ mapPacBio.sh ref=ref.hiv.fa in=SRR10971381_1.fq.gz in2=SRR10971381_2.fq.gz outm=hiv.mapped.sam vslow k=8 maxindel=0 minratio=0.1
$ samtools view hiv.mapped.sam|grep NM:i|wc -l # over 200,000 aligned reads that weren't multimapped so they weren't missing an NM:i tag
268534
$ samtools view hiv.mapped.sam|grep NM:i|cut -f6|ruby -ne'x=$_.scan(/\d+(?=[XID])/).map(&:to_i).sum;y=$_.scan(/\d+(?==)/).map(&:to_i).sum;puts [x*100.0/(x+y),$_]*" "'|sort -n|head -n4
10.0 23S1X9=
15.384615384615385 10=2X1=18S
15.384615384615385 10=2X1=19S
15.384615384615385 22S1X9=1X2=
When I ran minimap2 and Bowtie2 with the default parameters, I got zero reads that aligned against the HIV reference:
minimap2 -a --sam-hit-only hiv.fa SRR10971381_[12].fq.gz
bowtie2-build ref.hiv.fa{,};bowtie2 -x ref.hiv.fa --no-unal -p4 -1 SRR10971381_1.fq.gz -2 SRR10971381_2.fq.gz
When I ran BBMap with the default parameters, I got five reads that aligned against HIV, but the reads were only 31 to 36 bases long and each read had 7 to 9 errors:
$ bbmap.sh ref=ref.hiv.fa in=SRR10971381_1.fq.gz in2=SRR10971381_2.fq.gz outm=bbdefault.hiv.sam [...] $ grep NM:i: bbdefault.hiv.sam|cut -f1,6,10,12|sed 's/ [^\t]*//'|column -t SRR10971381.3293584 4X2=1X5=1X2=1X11=1X7=1X GTGGGAGGGAGACGGGGGAGAGAGAGAGAGAGACAG NM:i:9 SRR10971381.4837603 2X3=4X2=1X4=1X14= ATAAGACAGAGATGGACAGAGAGACAGAGAC NM:i:8 SRR10971381.10493971 2X3=4X2=1X4=1X14= ATAAGACAGAGATGGACAGAGAGACAGAGAC NM:i:8 SRR10971381.13295070 1X1=2X17=1X1=4X4= ATTCAGAGAGAGACAGAGACAGAGAGAAACA NM:i:8 SRR10971381.17916482 1=3X4=1I2=3X17= GGCAGAGACAGCCAGAGAGAGAGACAGAGAC NM:i:7
Mike Wallach wrote: "Eighth, he sought to discover whether one could take the sample and 'find' other claimed viruses in it. He searched for 'Hepatitus' and 'HIV' and found BOTH of them to have lower error rates than 'SARS-CoV-2.'" [https://theviraldelusion.substack.com/p/revealed-the-sars-cov-2-sequencing] He probably referred to this part of the paper: "Already in the previous section, a high structural similarity of the published sequences with the reference sequence LC312715.1 [HIV] was shown. The calculated consensus sequence showed relatively lower error rates R1 = 8.60% and R2 = 8.83% compared to e.g. the SARS associated references." (R1 means reads from the file for forward reads and R2 means reverse reads.)
Based on the code on page 23 of the paper, I think the mathematician may have first aligned Wu et al.'s reads against an HIV reference, and then he created a consensus sequence of the aligned reads by picking the most common nucleotide value at each position of the reference, he calculated the percentage of mismatched bases when he compared the consensus sequence against his HIV reference:

When I tried running code similar to the code above, I got an error rate of about 9.4% which is close enough to his figures (and there's a couple of different ways to calculate the error rate, and I don't know if he calculated the average of the error rates of individual reads and not total mismatches divided by total bases mapped like me):
$ fastp -i SRR10971381_1.fastq.gz -I SRR10971381_2.fastq.gz -o SRR10971381_1.fq.gz -O SRR10971381_2.fq.gz $ wget https://raw.githubusercontent.com/lh3/samtools/master/bcftools/vcfutils.pl -P ~/bin $ printf %s\\n MN908947.3\ sars2 LC312715.1\ hiv NC_001653.2\ hepatitis AF266291.1\ measles>refs $ cat refs|while read l m;do curl -s "https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=$l"|cut -d\ -f1 >ref.$y.fa;done $ mapPacBio.sh ref=ref.hiv.fa in=SRR10971381_1.fq.gz in2=SRR10971381_2.fq.gz outm=hiv.mapped.sam $ reformat.sh in=hiv.mapped.sam out=hiv.filtered.sam minlength=37 idfilter=60 ow=t $ samtools sort -@3 hiv.filtered.sam|bcftools mpileup -f ref.hiv.fa -|bcftools call -c|perl ~/bin/vcfutils.pl vcf2fq|seqtk seq -aQ64 -q20 -n N>hiv.cons.fa $ bwa index ref.hiv.fa;bwa mem -t4 ref.hiv.fa hiv.cons.fa|samtools stat|grep error [...] SN error rate: 9.446402e-02 # mismatches / bases mapped (cigar)
However when I repeated the same procedure for SARS2, my error rate was only about 3e-5. So I don't know what the mathematician meant by "relatively lower error rates" compared to the SARS-associated references.
And actually even in the case of HIV, all the errors were just spots
where the consensus sequence had an N base. When I removed the
seqtk command which replaced bases with quality below 20
with N letters, I now got zero mismatches for the HIV consensus:
$ samtools sort -@3 hiv.filtered.sam|bcftools mpileup -f ref.hiv.fa -|bcftools call -c|perl ~/bin/vcfutils.pl vcf2fq>hiv.cons.fq $ bwa mem -t4 ref.hiv.fa hiv.cons2.fa|samtools stat|grep error [...] SN error rate: 0.000000e+00 # mismatches / bases mapped (cigar) $ seqkit subseq -r1:50 hiv.cons.fa # low-quality bases were replaced with N letters in the file produced by the previous code block >LC312715.1 NNNNagNgcgcgcNgcaagaggcgaggggcggcaacNggtgagtacgccg $ seqkit subseq -r1:50 hiv.cons.fq # now there's no N letters when the `seqtk` command was removed @LC312715.1 ctgaagcgcgcgcagcaagaggcgaggggcggcaactggtgagtacgccg + ?QSHpvH~V~~~~Do]X~~~~~~~~~~~~~~~Vf~~N~~~~~n~h|k|wb
The mathematician from Hamburg also wrote: "With the sequence data at hand, we were able to compute consensus sequences for the reference genomes LC312715.1 (HIV) and NC_001653.2 (Hepatitis delta virus) with higher goodness than for those reference sequences we considered associated with coronaviruses. This was particularly true for bat-SL-CoVZC45 (GenBank: MG772933.1), which led to the origin hypothesis of SARS-CoV-2. Thus, we were able to substantiate our hypothesis that the claimed viral genome sequences are misinterpretations in the sense that they have been or are being constructed unnoticed from non-viral nucleic acid fragments." Table 4 of the supplementary PDF shows that the error rate for ZC45 was 10.2% for both forward and reverse reads: [https://impfen-nein-danke.de/u/structural_analysis_of_sequence_data_in_virology_tables_and_figures.pdf]

When I repeated the procedure above with ZC45, it got an error rate
of about 14% when I masked low-quality bases in the consensus with
N letters or about 11% when I didn't:
$ curl -s 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nucleotide&rettype=fasta&id=MG772933.1'|cut -d\ -f1 >ref.zc45.fa [...] $ bwa index ref.zc45.fa;bwa mem -t4 ref.zc45.fa zc45.cons.fa|samtools stat|grep error # masked with N letters [...] SN error rate: 1.363941e-01 # mismatches / bases mapped (cigar) [...] $ bwa mem -t4 ref.zc45.fa zc45.cons.fq|samtools stat|grep error # not masked SN error rate: 1.057312e-01 # mismatches / bases mapped (cigar)
The ZC45 consensus had N bases but the HIV consensus
didn't, which might partially explain why the ZC45 consensus had a
higher error rate. The reads that aligned against HIV were short and
they didn't actually come from HIV, so the errors in the reads may have
been random enough that they ended up canceling each other out in the
consensus sequence. But in the case of ZC45 many of the aligned reads
actually came from SARS2, so they had a systematic profile of mismatches
at the spots where SARS2 was different from ZC45 so the mismatches
didn't get canceled out. The ZC45 consensus actually got a lower error
rate when I aligned it against the SARS2 reference instead of the ZC45
reference:
$ bwa mem -t4 ref.sars2.fa zc45.cons.fq|samtools stat|grep error [...] SN error rate: 5.295835e-02 # mismatches / bases mapped (cigar)
If the soft-clipped parts of reads are removed, then the average length of the reads that aligned against ZC45 was about 144 bases, but the average length of the reads that aligned against HIV was only about 40 bases:
$ awk 'NR==FNR{if($6!="*")a[$1]=$6;next}$6=="*"{$6=a[$1]}1' {O,}FS=\\t zc45.filtered.sam{,}|cut -f6|sed 's/[0-9]*S//'|awk -F'[^0-9]+' '{s=0;for(i=1;i<=NF;i++)s+=$i;x+=s}END{print x/NR}'
144.244
$ awk 'NR==FNR{if($6!="*")a[$1]=$6;next}$6=="*"{$6=a[$1]}1' {O,}FS=\\t hiv.filtered.sam{,}|cut -f6|sed 's/[0-9]*S//'|awk -F'[^0-9]+' '{s=0;for(i=1;i<=NF;i++)s+=$i;x+=s}END{print x/NR}'
39.8861
And if you look at the aligned reads and not the consensus sequence based on the aligned reads, ZC45 has an error rate of about 13% but HIV has an error rate of about 39%:
$ samtools sort -@3 zc45.filtered.sam|samtools stats|grep error SN error rate: 1.255912e-01 # mismatches / bases mapped (cigar) $ samtools sort -@3 hiv.filtered.sam|samtools stats|grep error SN error rate: 3.881330e-01 # mismatches / bases mapped (cigar)
In October 2023, Tom Cowan published a video where he claimed that because the longest contig generated by the mathematician from Hamburg was not identical to the longest contig reported by Wu et al., it meant that Wu et al. manually manipulated their contig so they added in features which made the genome seem like it had been tinkered in a lab, and therefore they managed to trick people into believing in the lab leak narrative. [https://www.bitchute.com/video/Jt7uiAp9LfWs/] So Cowan said that therefore the paper by Wu et al. laid the foundation for the lab leak narrative.
Cowan said: "The Fan Wu people made this whole 30,000-string nucleotide up and specifically put in parts that would guarantee there would be this lab leak story, then that is the origin of the story. [...] With those 56 million plus pieces, can you actually assemble this 30,000 base pair genome, or did they simply make it up from scratch - not from scratch, they took what they got and then manually - in other words, these Fan Wu group put on the dodo bird part deliberately and specifically, and it was not actually in the original reads." [time 32:50] And a bit later Cowan said: "So us - meaning Stefan and our group that helped fund this - hired a mathematician who could do the sequencing, and said, 'Here, take these 56 million reads and see if you can get this 30,000 base pair genome.' And surprise surprise, he can't. That genome doesn't exist out of that raw data, which can only mean, and I mean only mean, that the Fan Wu group made that genome up based on - they took some 26, 28 thousand, 25 thousand - we don't know - and then actually added on manually pieces and specifically made them look like flesh-eating dodo bird beaks. In other words, they put, quote, 'scary spike protein' and 'cleavage site' sequences from other published sequences like HIV, and put those not into a particle, not into a virus that can be released, but into the computer database. That is the only place that genome exists, not in the real world." [time 40:00] (I didn't even know that the mathematician from Hamburg was hired by Lanka's group. Maybe he should've disclosed it as a conflict of interest.)
I tried running fastp and MEGAHIT with the same
parameters that were listed in the paper by the mathematician from
Hamburg:
curl "https://www.ebi.ac.uk/ena/portal/api/filereport?accession=SRR10971381&result=read_run&fields=fastq_ftp"| sed 1d|cut -f2|tr \; \\n|sed s,^,ftp://,|xargs wget -q fastp -i SRR10971381_1.fastq.gz -I SRR10971381_2.fastq.gz -o SRR10971381_1.fastq -O SRR10971381_2.fastq megahit -1 SRR10971381_1.fastq -2 SRR10971381_2.fastq -o megahit_result
My longest contig was 29,802 bases long like in the case of the mathematician from Hamburg, so it's probably identical to his contig. When I compared my contig to the first 30,474-base version of Wuhan-Hu-1 at GenBank, there was one nucleotide change in the nucleocapsid gene at position 29,293 of the alignment, but all other differences were in the untranslated regions at the beginning and end of the genome (so there were no differences in the regions of the spike protein which contain the HIV-like inserts or the furin cleavage site):
$ curl -s 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=MN908947.1'>wu1.fa
$ seqkit sort -lr megahit_result/final.contigs.fa|seqkit head -n1|seqkit seq -rp|cat - wu1.fa|mafft --thread 4 ->temp.aln
[...]
$ aldi2()(cat "$@">/tmp/aldi;Rscript --no-init-file -e 'm=t(as.matrix(Biostrings::readDNAStringSet("/tmp/aldi")));diff=which(rowSums(m!=m[,1])>0);split(diff,cumsum(diff(c(-Inf,diff))!=1))|>sapply(\(x)c(if(length(x)==1)x else paste0(x[1],"-",tail(x,1)),apply(m[x,,drop=F],2,paste,collapse="")))|>t()|>write.table(quote=F,sep=";",row.names=F)')
$ R -e 'if(!require("BiocManager"))install.packages("BiocManager");BiocManager::install("Biostrings")'
[...]
$ aldi2 temp.aln
;k141_7732 flag=1 multi=445.0000 len=29802;MN908947.1 Wuhan seafood market pneumonia virus isolate Wuhan-Hu-1, complete genome
1-11;----------G;CGGTGACGCAT
13-14;TT;CA
18-19;GG;CA
22-27;-----T;CCCACC
29293;G;C
29818-30473;--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------;CCTAATGTGTAAAATTAATTTTAGTAGTGCTATCCCCATGTGATTTTAATAGCTTCTTCCTGGGATCTTGATTTCAACAGCCTCTTCCTCATACTATTCTCAACACTACTGTCAGTGAACTTCTAAAAATGGATTCTGTGTTTGACCGTAGGAAACATTTCAGTATTCCTTATCTTTAGAAACCTCTGCAACTCAAGTGTCTCTGCCAAAGAGTCTTCAAGGTAATGTCAAATACCATAACATATTTTTTAAGTTTTTGTATACTTCCTAGGAATATATGTCATAGTATGTAAAGAAGATCTGATTCATGTGATCTGTACTGTTAAGAAACACTTCAAAGGAAAACAACCTCTCATAGATCTTCAAATCCTGTTGTTCTTGAGTTGGAATGTACAATATTCAGGTAGAGATGCTGGTACAAAAAAAGACTGAATCATCAGGCTATTAGAAAGATAAACTAGGCCTCTAACATAGTAACTTTAACAGTTTGTACTTATACATATTTTCACATTGAAATATAGTTTTATTCATGACTTTTTTTGTTTTAGCTTCTCTGTCTTCCATTATTTCAAGCTGCTAAAAATTAAAAATATCCTATAGCAAAGGGCTATGGCATCTTTTTGTAAAAATAAGGAAAGCAAGGTTTTTTGATAATC
The output above shows all segments of the alignment where my longest contig differs from the first version of Wuhan-Hu-1, where the first column shows the position of the segment, the second column shows the segment in my longest contig, and the third column shows the segment in the first version of Wuhan-Hu-1. From the last line of the output above, you can see that the first version of Wuhan-Hu-1 ends with a long piece of the human genome but my longest contig only has gaps at the corresponding region of the alignment. You can try copying the nucleotide sequence from the last line above, and then paste it to the big text box here and press BLAST: https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn. It shows that the sequence has 99.7% match to a human genome with about 93% query coverage (where there's not 100% coverage because BLAST skipped the first 38 bases of the segment because they came from the genome of SARS2 but they were missing from my contig).
Mark Bailey's article titled "Farewell to Virology" is divided into three parts, where the second part is about genetic sequencing and PCR. In a video about the second part that was done by Steve Falconer from Spacebusters, he asked: "If the virus came from a bat first and later jumped to humans, then how is its genomic sequence made out of common human sequences, if the virus isn't human and didn't even come from humans? Obviously because Fan Wu's team assembled a fake virus genome out of chopped up bits including the human lung sputum sample. It's a smoking gun." [https://www.bitchute.com/video/p1bkp55x78aK/, time 1:48:20] Then he showed a part of the paper by the mathematician from Hamburg which said: "At this point, the contig with the identification k141_27232, with which 1,407,705 sequences are associated, and thus about 5% of the remaining 26,108,482 sequences, should be discussed in detail. Alignment with the nucleotide database on 05/12/2021 showed a high match (98.85%) with 'Homo sapiens RNA, 45S pre-ribosomal N4 (RNA45SN4), ribosomal RNA' (GenBank: NR_146117.1, dated 04/07/2020). This observation contradicts the claim in [1] that ribosomal RNA depletion was performed and human sequence reads were filtered using the human reference genome (human release 32, GRCh38.p13)." And then Falconer asked: "If you filter out the human reads, then why are they still in your virus genome?"
However the part of the paper he quoted wasn't talking about the contig for SARS2, but about another contig assembled by MEGAHIT which consisted of a part of the human genome. In the supplementary PDF published by the mathematician from Hamburg, you can see the best BLAST hits for the contig if you search for "k141_27232" (or look at rows 12-16 of the table in the image below). [https://impfen-nein-danke.de/u/structural_analysis_of_sequence_data_in_virology_tables_and_figures.pdf]
When the mathematician from Hamburg wrote that "human sequence reads were filtered using the human reference genome", he referred to a part of the paper by Wu et al. which said: "To identify possible aetiological agents present in the sequencing data, the abundance of the assembled contigs was first evaluated as the expected counts using the RSEM program3[4] implemented in Trinity. Non-human reads (23,712,657 reads), generated by filtering host reads using the human genome (human release 32, GRCh38.p13, downloaded from Gencode) by Bowtie[235], were used for the RSEM abundance assessment." However that paragraph came after a part of the paper where they decribed running MEGAHIT on the original unfiltered reads, so Wu et al. probably filtered out human reads before they did the RSEM abundance assessment but not before they ran MEGAHIT. In the set of reads that Wu et al. uploaded to the NCBI's sequence read archive, they included all 56,565,928 reads but most human reads were masked so that they consisted of only N bases. But some human reads were missed by the masking procedure, which explains why the mathematician from Hamburg got some human contigs when he ran MEGAHIT on the reads from the SRA.
At time 1:49:55 in the video, Falconer said: "In their independent study, virologist Dr. Stefan Lanka and his mathematician took the original published genomic sequence data for the original work on coronavirus SARS-CoV-2 from Fan Wu's team - remember, based on the lung sputum fluid from a single patient in Wuhan, said to have only an 89.1% match to a group of SARS-like beta coronaviruses alleged to have been previously found in bats in China - and started running this one patient's sequences in GenBank against other available reference sequences - for not only things like bat SARS-CoV, but also other alleged virus genomes that have nothing to do with SARS, like HIV, hepatitis delta virus, measles virus, Zika virus, Ebola, and Marburg virus. And lo and behold, they were able to get anywhere from 82 percent to predominantly 96 to a 100 percent matches to all of these other viruses from the same one patient's sample sequences, much more than Fan Wu's 89% match to an alleged bat coronavirus."
However he mixed up two different parts of the paper, because the table with the 96-100% matches showed the BLAST hits for the 50 longest MEGAHIT contigs that were assembled by the mathematician from Hamburg. Almost all of the other contigs matched parts of the human genome or bacterial genomes, apart from one contig which matched SARS2, a few contigs with no matches, and one contig where the best match was titled "Reagent-associated CRESS-like virus 1 isolate 7 replicase-like gene, partial sequence". But the table didn't include HIV or hepatitis delta which were discussed in a different part of the paper. The reason why the table includes matches for SARS2, ZC45, and SARS1 is because it didn't just include the best match for the SARS2 contig but also matches to ZC45 and SARS1 for comparison, but from the table below you can see that the viruses on the first 5 rows all match the same contig: [https://impfen-nein-danke.de/u/structural_analysis_of_sequence_data_in_virology_tables_and_figures.pdf]

In June 2024 Mike Wallach published a Substack post about the paper by the mathematician from Hamburg. [https://theviraldelusion.substack.com/p/revealed-the-sars-cov-2-sequencing]
Wallach wrote that one part of the sequencing procedure was to "Submit this mixture to a PCR process to amplify the number of the RNA fragments, including the 'amplification' of any specific RNA fragments the virologists expect to find." However Wu et al. did metagenomic sequencing where they weren't using PCR to do targeted enrichment for any specific organism or genetic region, but they only did random priming as part of the procedure of converting RNA to cDNA.
Wallach wrote: "Fourth, he found that up to 17% of the final sequence was based on RNA contigs specifically targeted and then 'found' by the PCR process at cycle thresholds of ct 35 to 45, cycle counts well known in the literature to 'find' anything you want." I believe he referred to this sentence: "While de novo meta-transcriptomic assembly often uses no reference sequences or only downstream reference sequences, whole genome sequencing uses a large number of specific primer sequences, some of which already together cover 4% to 17% of the target genome [1, 17]." However the PCR protocols he wrote about were published after Wu et al. had already done the de-novo assembly of the genome, so it doesn't make sense to say that the contigs Wu et al. assembled would've somehow been based on those PCR primers. And how would you even design PCR primers if you don't first know the sequence of the virus they're supposed to target?
Wallach wrote: "Sixth, he found that contigs in the remaining data sample AFTER the Fan Wu paper claims to have filtered it for known human RNA, matched known human RNA." However the only sentence where Wu et al. wrote about filtering human reads said: "Non-human reads (23,712,657 reads), generated by filtering host reads using the human genome (human release 32, GRCh38.p13, downloaded from Gencode) by Bowtie2[35], were used for the RSEM abundance assessment." So Wu et al. wrote that they filtered out human reads before they did RSEM abundance assessment, but not that they filtered out human reads from the set of reads they uploaded to the Sequence Read Archive. In the reads that were uploaded to the SRA, the vast majority of human reads had been masked with N bases for privacy reasons, but there's a small percentage of human reads that didn't get masked.
USMortality took the reference genomes of different viruses, and he
generated random short reads for each virus with the wgsim
utility which comes with SAMtools and with his own TypeScript script.
[https://usmortality.substack.com/p/how-reliable-is-megahit]
When he tried to assemble the short reads with MEGAHIT, he failed to get
a complete contig for some viruses like HKU1, porcine adenovirus, HIV-1,
and HIV-2:

From the table of percentages above, you can see that when
USMortality used wgsim to simulate short reads for the
reference genome of HKU1, his longest contig was only about 89% of the
length of the reference genome. I tried replicating his experiment so
that I used wgsim to generate 100,000 70-base reads for the
reference genome of HKU1. When I ran MEGAHIT to assemble the reads, I
got two different contigs, and the longer contig was only 26,533 bases
long even though the reference genome of HKU1 is 29,926 bases long:
$ brew install megahit -s # `-s` installs from source because the bottled version was compiled with GCC9 so I got an error when I tried to run it
[...]
$ brew install seqkit samtools
[...]
$ curl -s 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=NC_006577'>hku1.fa
$ wgsim -N100000 hku1.fa hku1_{1,2}.fq
[...]
$ megahit -1 hku1_1.fq -2 hku1_2.fq -o megahku
[...]
$ seqkit stat hku1.fa megahku/final.contigs.fa|column -t
file format type num_seqs sum_len min_len avg_len max_len
hku1.fa FASTA DNA 1 29,926 29,926 29,926 29,926
megahku/final.contigs.fa FASTA DNA 2 29,634 3,101 14,817 26,533
When I aligned the contigs with Bowtie2, I noticed that my two contigs covered positions 7-3108 and 3394-29927, so there was a gap of 285 bases between the contigs. But then I noticed that the gap fell within a region where the same 30-base segment was repeated 9 times, where there's also a few additional repeats which did not fall within the gap between the contigs:
$ bowtie2-build hku1.fa{,}
[...]
$ bowtie2 -p4 -x hku1.fa -fU megahku/final.contigs.fa|samtools sort ->hku1.bam
[...]
$ samtools view hku1.bam|awk -F\\t '{l=length($10);print$4,$6,l,$4+l}'|column -t
7 647M1I2453M 3101 3108
3394 26533M 26533 29927
$ seqkit subseq -r 3109:3393 hku1.fa
>NC_006577.2 Human coronavirus HKU1, complete genome
AGATGTTGTTACTGGTGACAATGACGATGAAGATGTTGTTACTGGTGACAATGACGATGA
AGATGTTGTTACTGGTGACAATGACGATGAAGATGTTGTTACTGGTGACAATGACGATGA
AGATGTTGTTACTGGTGACAATGACGATGAAGATGTTGTTACTGGTGACAATGACGATGA
AGATGTTGTTACTGGTGACAATGACGATGAAGATGTTGTTACTGGTGACAATGACGATGA
AGATGTTGTTACTGGTGACAATGACGATGAAGATGTTGTTACTGG
A paper about HKU1 said: "Genome analysis also
revealed various numbers of tandem copies of a perfect 30-base acidic
tandem repeat (ATR) which encodes NDDEDVVTGD and various numbers and
sequences of imperfect repeats in the N terminus of nsp3 inside the
acidic domain upstream of papain-like protease 1 among the 22
genomes."
[https://pubmed.ncbi.nlm.nih.gov/16809319/]
But NDDEDVVTGD is the translation of the 30-base repeated
segment:
seqkit translate<<<$'>a\nAATGACGATGAAGATGTTGTTACTGGTGAC'.
Above I ran wgsim using the default read length which is
70 bases, and I used paired reads instead of unpaired reads. But if
MEGAHIT would've had to assemble the contigs from unpaired reads that
each were 70 bases long, how could it know how many times the 30-base
segment was repeated if it can only see one a single 70-base window of
the genome at a time? Actually one of the main reasons why the paired
read layout is used is that it helps to assemble regions with repeats
because the read pair covers a region that is longer than an individual
read.
[https://emea.illumina.com/science/technology/next-generation-sequencing/plan-experiments/paired-end-vs-single-read.html]
But even though wgsim generated paired reads, I guess the
region with repeats was so long and the read length was so short that
the paired layout didn't help MEGAHIT to assemble the region correctly.
However I also tried using a longer read length with wgsim,
and I tried using a higher k-value with MEGAHIT, but neither helped with
getting a complete contig for HKU1.
USMortality also failed to assemble a complete contig for the reference genome of porcine adenovirus, but that's because it contains a tandem repeat where the same 724-base segment is repeated twice in a row near the 3' end:
$ curl 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=NC_044935'>adeno.fa
$ seqkit seq -s adeno.fa|awk '{for(i=1;;i++){delete a;for(j=1;j<=length-i+1;j++)a[substr($0,j,i)][j];found=0;for(x in a){if(length(a[x])>1){found=1;for(y in a[x])print i,y,y+length(x)-1,x}}if(!found)exit}}'|sort -srnk1,1|awk 'NR>1&&$1!=x{exit}{x=$1}1'
725 29481 30205 CGAAGGACAACTCACCGTCCCCGCCACGGCCCCTTTAGTCTCAGGGAGCGCAGGCATCTCTTTCAACTACTCCAGCAATGACTTCGTCTTAGACAATGACAGTCTCAGTTTGAGGCCAAAGGCCATCTCTGTCACCCCTCCGCTGCAGTCCACAGAGGACACAATCTCCCTGAATTATTCTAACGACTTTTCTGTGGACAATGGCGCCCTCACCTTGGCTCCAACTTTCAAACCCTACACGCTGTGGACTGGCGCCTCACCCACAGCAAATGTCATTCTAACAAACACCACCACTCCCAACGGCACCTTTTTCCTATGCCTGACACGTGTGGGTGGGTTAGTTTTGGGTTCCTTTGCCCTGAAATCATCCATCGACCTTACTAGTATGACCAAAAAGGTCAATTTTATTTTTGATGGGGCAGGTCGGCTTCAGTCAGACTCCACTTATAAAGGGAGATTTGGATTTAGATCCAACGACAGCGTAATTGAACCCACAGCCGCAGGACTCAGTCCAGCCTGGTTAATGCCAAGCACCTTTATTTATCCACGCAACACCTCCGGTTCTTCCCTAACATCATTTGTATACATTAATCAGACATATGTGCATGTGGACATCAAGGTAAACACACTCTCTACAAACGGATATAGCCTAGAATTTAACTTTCAAAACATGAGCTTCTCCGCCCCCTTCTCCACCTCCTACGGGACCTTCTGCTACGTGCCCC
725 30205 30929 CGAAGGACAACTCACCGTCCCCGCCACGGCCCCTTTAGTCTCAGGGAGCGCAGGCATCTCTTTCAACTACTCCAGCAATGACTTCGTCTTAGACAATGACAGTCTCAGTTTGAGGCCAAAGGCCATCTCTGTCACCCCTCCGCTGCAGTCCACAGAGGACACAATCTCCCTGAATTATTCTAACGACTTTTCTGTGGACAATGGCGCCCTCACCTTGGCTCCAACTTTCAAACCCTACACGCTGTGGACTGGCGCCTCACCCACAGCAAATGTCATTCTAACAAACACCACCACTCCCAACGGCACCTTTTTCCTATGCCTGACACGTGTGGGTGGGTTAGTTTTGGGTTCCTTTGCCCTGAAATCATCCATCGACCTTACTAGTATGACCAAAAAGGTCAATTTTATTTTTGATGGGGCAGGTCGGCTTCAGTCAGACTCCACTTATAAAGGGAGATTTGGATTTAGATCCAACGACAGCGTAATTGAACCCACAGCCGCAGGACTCAGTCCAGCCTGGTTAATGCCAAGCACCTTTATTTATCCACGCAACACCTCCGGTTCTTCCCTAACATCATTTGTATACATTAATCAGACATATGTGCATGTGGACATCAAGGTAAACACACTCTCTACAAACGGATATAGCCTAGAATTTAACTTTCAAAACATGAGCTTCTCCGCCCCCTTCTCCACCTCCTACGGGACCTTCTGCTACGTGCCCC
USMortality also failed to get complete contigs for HIV-1 and HIV-2, but that's because HIV has a long terminal repeat where a long segment at the 5' UTR is repeated at the 3' UTR: [https://en.wikipedia.org/wiki/Long_terminal_repeat]
The HIV-1 LTR is 634 bp[5] in length and, like other retroviral LTRs, is segmented into the U3, R, and U5 regions. U3 and U5 has been further subdivided according to transcription factor sites and their impact on LTR activity and viral gene expression. The multi-step process of reverse transcription results in the placement of two identical LTRs, each consisting of a U3, R, and U5 region, at either end of the proviral DNA. The ends of the LTRs subsequently participate in integration of the provirus into the host genome. Once the provirus has been integrated, the LTR on the 5' end serves as the promoter for the entire retroviral genome, while the LTR at the 3' end provides for nascent viral RNA polyadenylation and, in HIV-1, HIV-2, and SIV, encodes the accessory protein, Nef.[6]
The long terminal repeat is 855 bases long in the reference genome of HIV-2:
$ curl -s 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=NC_001722'>hiv2.fa $ seqkit subseq -r 1:855 hiv2.fa|seqkit locate -f- hiv2.fa|cut -f1,4-6|column -t seqID strand start end NC_001722.1 + 1 855 NC_001722.1 + 9505 10359
In order to assemble regions with repeats in MEGAHIT, it sometimes helps to use a higher k-value, but it won't be very useful with these huge terminal repeats in HIV since the maximum k-mer value supported by MEGAHIT is only 255. The paper about MEGAHIT says: "While a small k-mer size is favourable for filtering erroneous edges and filling gaps in low-coverage regions, a large k-mer size is useful for resolving repeats." [https://academic.oup.com/bioinformatics/article/31/10/1674/177884] And the paper about SPAdes says: "The choice of k affects the construction of the de Bruijn graph. Smaller values of k collapse more repeats together, making the graph more tangled. Larger values of k may fail to detect overlaps between reads, particularly in low coverage regions, making the graph more fragmented. Since low coverage regions are typical for SCS data, the choice of k greatly affects the quality of single-cell assembly. Ideally, one should use smaller values of k in low-coverage regions (to reduce fragmentation) and larger values of k in high-coverage regions (to reduce repeat collapsing)." [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3342519/]
USMortality wrote that when he mixed together simulated reads from multiple different viruses into a single FASTA file and he assembled the reads with MEGAHIT, his longest contigs for both SARS2 and SARS1 only covered about 51% of the reference genome: [https://usmortality.substack.com/p/how-reliable-is-megahit]

When I used wgsim to simulate reads for SARS2 and SARS1
and I ran MEGAHIT on the concatenated reads, I got three contigs for
both SARS1 and SARS2. I got one contig for SARS1 which covered positions
1-15046 and another contig which covered positions 14992-29707, so for
some reason the contigs were not merged even though they had 47
overlapping bases, and I got similar overlapping contigs for SARS2 which
were split around position 15,100:
$ curl -s 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=NC_004718.3'>sars1.fa
$ curl -s 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=MN908947.3'>sars2.fa
$ for i in 1 2;do wgsim -N100000 sars$i.fa sars$i\_{1,2}.fq;done;for i in 1 2;do cat sars[12]_$i.fq>sars3_$i.fq;done
[...]
$ megahit -1 sars3_1.fq -2 sars3_2.fq -o megasars3
[...]
$ cat sars[12].fa>sars12.fa
$ bowtie2-build sars12.fa{,};bowtie2 -x sars12.fa -fU megasars3/final.contigs.fa|samtools sort ->sars3.bam
[...]
$ samtools view sars3.bam|awk -F\\t '{print$1,$3,$4,$6,length($10)}'|column -t
k79_7 NC_004718.3 1 15045M 15045
k79_6 NC_004718.3 14992 14714M 14714
k79_3 NC_004718.3 25165 247M 247
k79_5 MN908947.3 10 8109M1D6598M1D397M 15104
k79_0 MN908947.3 15062 2787M 2787
k79_2 MN908947.3 17779 64M1I11321M2D552M3D128M 12066
Then when I tried comparing parts of the SARS1 and SARS2 genomes around the region where the contigs were split, I noticed that SARS2 and SARS1 had an identical 74-base segment at that region:
$ (seqkit subseq -r 14950:15100 sars1.fa;seqkit subseq -r 15000:15200 sars2.fa)|mafft --clustalout -
[...]
NC_004718.3 --------------------ttttcgcgtatactaagcgtaatgtcatccctactataac
MN908947.3 ttatgaggatcaagatgcacttttcgcatatacaaaacgtaatgtcatccctactataac
*******.***** **.***********************
NC_004718.3 tcaaatgaatcttaagtatgccattagtgcaaagaatagagctcgcaccgtagctggtgt
MN908947.3 tcaaatgaatcttaagtatgccattagtgcaaagaatagagctcgcaccgtagctggtgt
************************************************************
NC_004718.3 ctctatctgtagtactatgacaaatagacagtttcatcagaaattattgaa---------
MN908947.3 ctctatctgtagtactatgaccaatagacagtttcatcaaaaattattgaaatcaatagc
********************* *****************.***********
NC_004718.3 ---------------------
MN908947.3 cgccactagaggagctactgt
The default read length used by wgsim is 70 bases, which is a few bases shorter than the length of the shared segment, but I failed to get single contigs for SARS1 and SARS2 even after I tried increasing the read length to 150 or 300. However when I increased the maximum k-value of MEGAHIT from 141 to 161, I was able to get single contigs for both SARS1 and SARS2 even with the default read length of 70:
$ megahit -1 sars3_1.fq -2 sars3_2.fq -o megasars4 --k-list 21,29,39,59,79,99,119,141,161 [...] 2023-10-27 15:43:10 - 2 contigs, total 59634 bp, min 29751 bp, max 29883 bp, avg 29817 bp, N50 29883 bp 2023-10-27 15:43:10 - ALL DONE. Time elapsed: 47.084499 seconds
USMortality downloaded the raw reads from a paper from 2020 where the genome of SARS2 was sequenced with direct RNA nanopore sequencing. [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7179501/] He then aligned the reads against the reference genome of SARS2, and he posted the screenshots from IGV below with the description: "As you can see, the quality of these long Nanopore reads is really terrible - so many errors and inserts! I could not find a single read that perfectly aligns to either the head or the tail! Note, that the highlighted letters are errors, and the purple/black areas are inserts/deletions. Judge for yourself, but in my opinion, the amount of errors, does not increase confidence in the genome." [https://usmortality.substack.com/p/why-do-wu-et-al-2020-refuse-to-answer]
The reason why the reads in his screenshots are full of N bases and they have no T bases is that USMortality forgot to convert U bases to T bases. And the reason why most reads appear to be full of errors is that they are actually soft-clipped parts of reads which were shown by IGV because he had enabled the "Show soft-clipped bases" option from the "Alignments" tab of the preferences. By default IGV doesn't display all letters within reads but only letters for bases which have a mismatch from the reference, but IGV displays all letters within soft-clipped parts of reads which makes them look like mismatches (and the UI of IGV is a bit confusing because there is no visual indication of which parts of reads are soft-clipped).
In order to accurately gauge the number of mismatches in IGV, you also have to uncheck "Quick consensus mode" from the context menu of the alignment view, because otherwise IGV will display mismatches as blanks unless the percentage of mismatches at the reference position is above a given threshold. [https://www.pacb.com/blog/igv-3-improves-support-pacbio-long-reads/] In order to save memory, IGV only displays up to about 100 reads for each position of the reference genome by default, but you can show all reads by unchecking "Preferences > Alignments >"Downsample reads". [https://software.broadinstitute.org/software/igv/book/export/html/37]
I downloaded the same nanopore reads that USMortality aligned in his
post. [https://osf.io/8f6n9, file
NanoporeDRS/Vero-Infected/20200222_0650_MN26136_FAN04901_ada1e2bf/VeroInf24h.all.fastq.zst]
I then aligned the reads with minimap2:
curl -s 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=MN908947.3'>sars2.fa minimap2 -a --sam-hit-only -x map-ont sars2.fa <(seqkit seq --rna2dna Downloads/VeroInf24h.all.fastq.zst)>nano7.sam;samtools sort nano7.sam>nano7.bam;samtools index nano7.bam
When I opened the aligned reads in IGV, I disabled the option to show soft-clipped parts of reads, I disabled the downsampling option, and I disabled the quick consensus mode, I got 9 reads with starting position 1, and all except one of the reads had zero mismatches from Wuhan-Hu-1 within the first 10 bases (even though all reads had over a thousand hard-clipped bases at the start which are not shown below by IGV):
Added later: USMortality later edited the Substack post to fix his screenshots. But he now said it was somehow problematic how a large proportion of the reads had clipped bases at either end. But I pointed out that chimeric reads are a well-known phenomenon, and if they would somehow be an insurmountable problem then people could use other sequencing methods or other library preparation methods that don't result in chimeric reads. When I asked ChatGPT why direct RNA nanopore sequencing would result in chimeric reads, it answered:
1. Template Switching During Reverse Transcription (if applicable): In some workflows, RNA molecules are reverse transcribed before sequencing. The reverse transcriptase enzyme can occasionally switch between different RNA templates, leading to the creation of chimeric sequences.
2. RNA Fragment Annealing or Ligation: During library preparation, RNA fragments from different molecules may anneal or be ligated together. If these chimeric RNA molecules are then sequenced, the resulting reads will appear as chimeras.
3. Nanopore Detection Errors: Nanopore sequencing directly detects RNA molecules passing through a nanopore. If two RNA molecules simultaneously enter or transiently interact within the pore, the electrical signal may become a composite, leading to a chimeric interpretation during basecalling.
4. Molecular Crowding or Sticky RNA Regions: RNA molecules with sticky sequences, such as homopolymers or repetitive regions, may transiently bind to other molecules. This interaction can result in misleading signals interpreted as a single continuous molecule.
Then USMortality replied: "Additionally, this phenomenon should only occur in a subset of reads. If it is observed consistently across all reads, it is more likely to reflect an actual biological feature rather than an artifact, such as chimeric reads." [https://x.com/USMortality/status/1866792958478995927]
But I told him that the phenomenon of chimeric reads does occur in only a subset of reads. I got 8,773 reads that didn't have any clipped bases at the start or end:
$ minimap2 -aY --sam-hit-only -t4 sars2.fa VeroInf24h.all.fq|samtools sort -@3 ->VeroInf24h.all.bam $ samtools view VeroInf24h.all.bam|awk '$6!~/S/'|wc -l 8773
The longest one of the reads that didn't have any clipped bases at the ends was 29,453 bases long. It had more deletions than insertions relative to Wuhan-Hu-1, so it was only missing the first 11 and last 23 bases of Wuhan-Hu-1:
$ samtools view VeroInf24h.all.bam|awk '$6!~/S/'|samtools fasta|seqkit sort -lr|seqkit head -n1|cat - sars2.fa|mafft --thread 4 --clustalout -
6bde649c-c7a2-4 -----------atacc-tcccaggtaacaaaccaaccatattttcgattcttgtagatct
MN908947.3 attaaaggtttataccttcccaggtaacaaaccaaccaactttcgatctcttgtagatct
***** ********************* ***. . .************
[...]
6bde649c-c7a2-4 caaattaatttagtg-tatccccatgtgattttaatagctt-ttaggagaatgacaaaaa
MN908947.3 ttaattttagtagtgctatccccatgtgattttaatagcttcttaggagaatgacaaaaa
. **** ***** ************************* ******************
6bde649c-c7a2-4 aaaaa-----------------------
MN908947.3 aaaaaaaaaaaaaaaaaaaaaaaaaaaa
In all of the longest reads without clipped bases, there were at least 11 bases missing at the 5' end. The paper where the reads were from said: "The SARS-CoV-2 genome was almost fully covered, missing only 12 nt from the 5' end due to the known inability of DRS to sequence the terminal ∼12 nt (Figure 2B)." [https://pmc.ncbi.nlm.nih.gov/articles/PMC7179501/] ChatGPT said:
The inability of direct RNA sequencing (DRS) to sequence the terminal ~12 nucleotides of RNA molecules, such as the SARS-CoV-2 genome, stems from technical and physical limitations in the nanopore sequencing process:
1. Nanopore Structure and Motor Protein Limitations: During DRS, a motor protein unwinds and feeds the RNA strand through the nanopore for sequencing. The motor protein must remain bound to the RNA to control its movement. As the RNA approaches its 5' end, the motor protein cannot maintain its grip on the molecule, leaving the terminal nucleotides unsequenced.
2. RNA Length Relative to Adapter: The RNA molecule is attached to an adapter that facilitates the interaction with the sequencing pore. The adapter prevents complete threading of the RNA, leaving the last few nucleotides unreachable by the nanopore for signal detection.
3. Signal Resolution and Decay: The electrical signal produced by the nanopore becomes less reliable as the RNA molecule shortens, leading to poor resolution or complete loss of signal for the terminal nucleotides.
USMortality wrote: "Even when trimming off the leading three or four nucleotides from the reads, Megahit still assembles de-novo the head with two extra G bases at the head!" [https://usmortality.substack.com/p/why-do-wu-et-al-2020-refuse-to-answer]
However I was able to get rid of the two extra G bases when I trimmed
the reads with fastp -f 5 -t 5, which removes the first 5
and last 5 bases from both forward and reverse reads. Then my longest
MEGAHIT contig was 29,870 bases long, and it was otherwise identical to
Wuhan-Hu-1 except it was missing the entire 33-base poly(A) tail.
There's 25 forward reads which match the first 20 bases of Wuhan-Hu-1 but which have two extra G bases added to the start:
$ seqkit locate -p GGATTAAAGGTTTATACCTTCC SRR10971381_1.fastq|cut -f1,4-|column -t seqID strand start end matched SRR10971381.718581 - 119 140 GGATTAAAGGTTTATACCTTCC SRR10971381.758993 - 86 107 GGATTAAAGGTTTATACCTTCC SRR10971381.1921582 - 105 126 GGATTAAAGGTTTATACCTTCC SRR10971381.2004837 - 105 126 GGATTAAAGGTTTATACCTTCC SRR10971381.2090745 - 105 126 GGATTAAAGGTTTATACCTTCC SRR10971381.2211935 - 105 126 GGATTAAAGGTTTATACCTTCC SRR10971381.2727470 - 105 126 GGATTAAAGGTTTATACCTTCC SRR10971381.4673132 - 103 124 GGATTAAAGGTTTATACCTTCC SRR10971381.6216519 - 104 125 GGATTAAAGGTTTATACCTTCC SRR10971381.6675729 - 117 138 GGATTAAAGGTTTATACCTTCC SRR10971381.7692719 - 105 126 GGATTAAAGGTTTATACCTTCC SRR10971381.8056736 + 3 24 GGATTAAAGGTTTATACCTTCC SRR10971381.10755198 - 105 126 GGATTAAAGGTTTATACCTTCC SRR10971381.11089222 - 105 126 GGATTAAAGGTTTATACCTTCC SRR10971381.12331153 - 105 126 GGATTAAAGGTTTATACCTTCC SRR10971381.17552714 - 105 126 GGATTAAAGGTTTATACCTTCC SRR10971381.19351707 - 120 141 GGATTAAAGGTTTATACCTTCC SRR10971381.19590294 - 104 125 GGATTAAAGGTTTATACCTTCC SRR10971381.19788065 - 104 125 GGATTAAAGGTTTATACCTTCC SRR10971381.22231571 - 117 138 GGATTAAAGGTTTATACCTTCC SRR10971381.23063694 - 105 126 GGATTAAAGGTTTATACCTTCC SRR10971381.23201569 - 105 126 GGATTAAAGGTTTATACCTTCC SRR10971381.24938341 - 119 140 GGATTAAAGGTTTATACCTTCC SRR10971381.27644056 - 119 140 GGATTAAAGGTTTATACCTTCC SRR10971381.28160997 - 86 107 GGATTAAAGGTTTATACCTTCC
In all of the reads shown above except a single read, the match is on the minus strand so the two extra G bases at the start of the read are actually two extra C bases at the end of the read. So that's why you have to trim the 3' ends of the reads in order to get rid of the extra G bases at the start of the contig.
The reverse reads are sense relative to DNA and therefore antisense relative to RNA, so they're what people might intuitively expect to be the forward reads. The manual of the Takara RNA-Seq kit used by Wu et al. says: "For sequencing libraries produced with this kit, Read 1 generates sequences antisense to the original RNA. This contrasts with libraries produced by the original SMARTer Stranded Total RNA-Seq Kit - Pico Input Mammalian, for which Read 1 generates sequences sense to the original RNA." [https://www.takarabio.com/documents/User+Manual/SMARTer+Stranded+Total+RNA/SMARTer+Stranded+Total+RNA-Seq+Kit+v2+-+Pico+Input+Mammalian+User+Manual_050619.pdf] An answer at BioStars said that the reads in the Read 1 file are called forward reads regardless of whether they are sense or antisense relative to the sequenced genome: "For example, with Illumina paired-end sequencing, you get two reads: commonly called 'forward' and 'reverse' reads, or even just 'Read 1' and 'Read 2'. In this case, the terms 'forward' and 'reverse' refer only to the relative orientation of each read to the other, and do not in themselves imply anything about their orientation relative to, say, the coding strand of the original RNA molecule in an RNA-seq experiment. When you do a strand-specific protocol, depending on the protocol, these reads will now have an assigned strandedness relative to the original RNA molecule. For example, a particular protocol might be designed that the reverse read just happens to represent the actual sequence of the RNA, while the forward read represents the reverse-complement sequence." [https://www.biostars.org/p/46769/#46975]
One reason why USMortality says that the genome of SARS-CoV-2 has not been validated is that MEGAHIT contigs for Wu et al.'s raw reads don't contain the full 33-base poly(A) tail that is included in the third version of Wuhan-Hu-1.
However the full poly(A) tail is not even included in Wu et al.'s raw reads. ChatGPT gave these reasons for why the reads might not include the full poly(A) tail:
- Fragmentation during Library Preparation: In metagenomic sequencing, the RNA or DNA is typically fragmented during the library preparation step to create a size distribution that is suitable for sequencing. This fragmentation process can result in the loss of the complete poly(A) tail, especially since the poly(A) tail is a homopolymeric sequence that may be more susceptible to shearing. [...]
- Primer Design and Amplification Bias: During reverse transcription (for RNA viruses), primers used to convert RNA into complementary DNA (cDNA) may not efficiently bind to the poly(A) tail, leading to incomplete reverse transcription of the tail. Additionally, amplification steps in library preparation can introduce bias where certain sequences are preferentially amplified over homopolymeric regions like poly(A) tails. [...]
- Biological Processing: In some cases, the poly(A) tail may be naturally shortened or modified during the viral life cycle or host processing of the viral RNA, resulting in variable lengths that may not always be fully captured in sequencing reads.
ChatGPT also said:
Yes, random priming used to convert RNA to cDNA can lead to the loss of the full poly(A) tail in an RNA virus. Here's why this happens:
- Random Primer Binding: Random primers bind to various locations along the RNA molecule, not specifically to the poly(A) tail. As a result, the cDNA synthesis initiated by these primers will start at random positions along the RNA. If a primer binds upstream of the poly(A) tail, the resulting cDNA will include only part of the poly(A) tail or none of it, depending on where the primer binds.
- Incomplete Coverage: Since random priming does not guarantee coverage of the very 3' end of the RNA where the poly(A) tail is located, many cDNA fragments generated will not extend into the poly(A) tail at all. This results in cDNA molecules that lack the full poly(A) sequence.
- Fragmentation and Size Selection: During library preparation, RNA is often fragmented to create smaller pieces suitable for sequencing. This fragmentation can occur before or after cDNA synthesis. If fragmentation occurs before reverse transcription, the poly(A) tail may be partially or entirely lost in the fragmentation process. If it occurs after, the resulting cDNA fragments may include only portions of the poly(A) tail.
- Bias Against Homopolymeric Sequences: Random primers may have a lower efficiency in binding to and initiating cDNA synthesis in homopolymeric regions like the poly(A) tail due to secondary structure formation or sequence composition. This can lead to a bias against the inclusion of the full poly(A) tail in the cDNA library.
- Subsequent Amplification Steps: PCR amplification of cDNA libraries can introduce biases where sequences that are easier to amplify (non-homopolymeric regions) are preferentially amplified over homopolymeric regions like the poly(A) tail. This can result in reduced representation of the poly(A) tail in the final sequencing library.
There is also a lot of variation in the length of the poly(A) tail throughout the life cycle of a virus, so it's not reasonable to require that the poly(A) tail should always be precisely 33 bases long: [https://www.cell.com/cell/fulltext/S0092-8674%2820%2930406-2]
We recently developed software to measure the length of poly(A) tail from DRS data (Y. Choi and H.C., unpublished data). Using this software, we confirm that, like other CoVs, SARS-CoV-2 RNAs carry poly(A) tails (Figures 4A-4B). The tail of viral RNAs is 47 nt in median length. The full-length gRNA has a relatively longer tail than sgRNAs. Notably, sgRNAs have two tail populations: a minor peak at ∼30 nt and a major peak at ∼45 nt (Figure 4B, arrowheads). Wu et al., 2013 previously observed that the poly(A) tail length of bovine CoV mRNAs changes during infection: from ∼45 nt immediately after virus entry to ∼65 nt at 6-9 hours post-infection and ∼30 nt at 120-144 hours post-infection. Thus, the short tails of ∼30 nt observed in this study may represent aged RNAs that are prone to decay.
One reason why USMortality says that the genome has not been validated is because when he aligns Wu et al.'s reads against Wuhan-Hu-1, he cannot find any read which has a perfect match to the 5' end, but there's always 3-5 extra bases at the start of reads that don't match Wuhan-Hu-1.
McKernan explained the purpose of the bases in this tweet: [https://x.com/Kevin_McKernan/status/1674577464184512513]
Do you know what random priming means? It means the hexamer that primes the second strand synthesis, is 5'-NNNNNN-3OH. The kit uses a strand flipping enzyme which adds bases on the watson strand of the 5' end. These become adaptors.
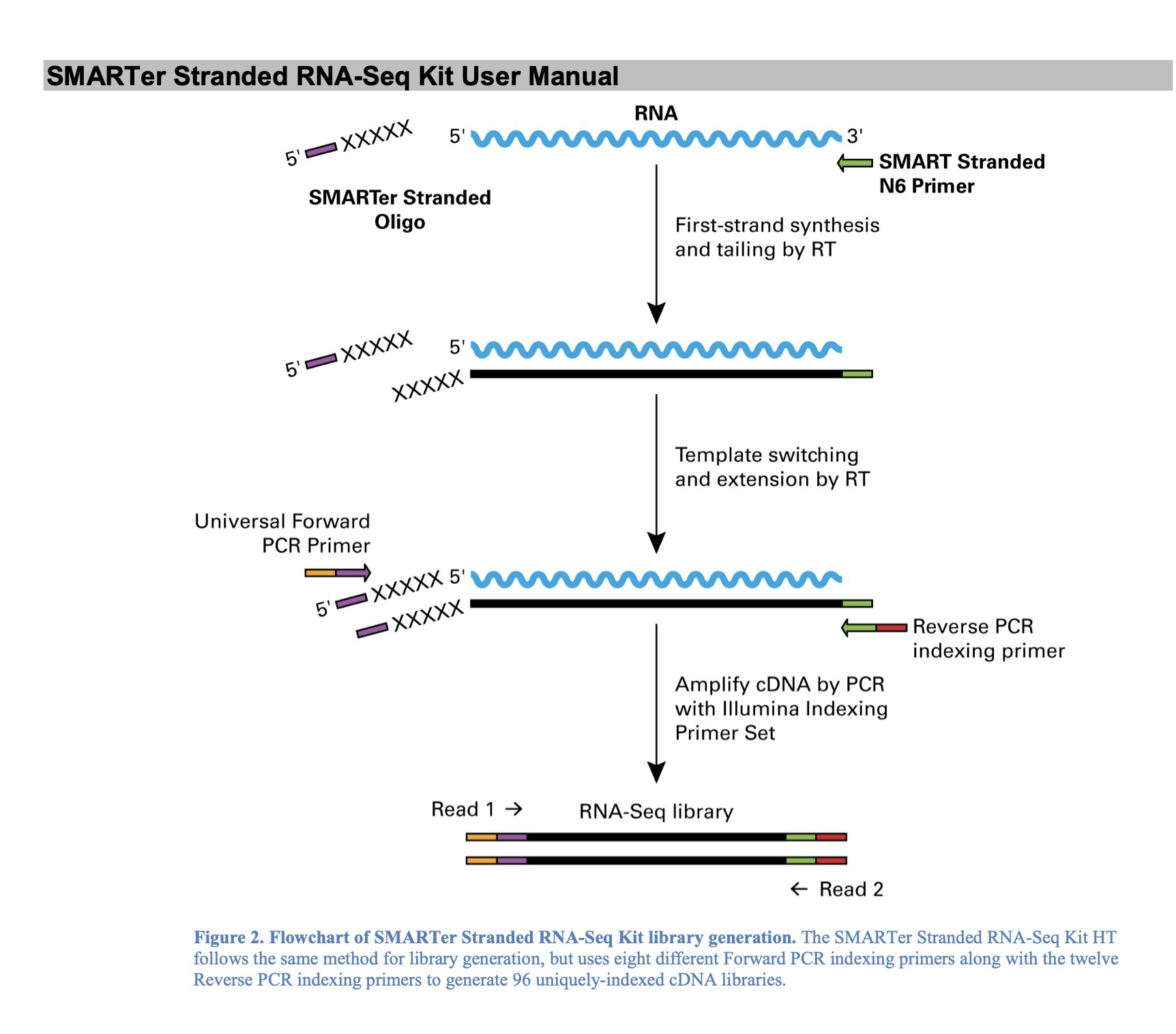
McKernan's screenshots are from the manual of the Takara SMARTer Stranded Total RNA-Seq kit v.2 that was used by Wu et al. [https://www.takarabio.com/documents/User+Manual/SMARTer+Stranded+Total+RNA/SMARTer+Stranded+Total+RNA-Seq+Kit+v2+-+Pico+Input+Mammalian+User+Manual_050619.pdf]
Actually the 5' template switching oligo is the key feature of the SMART method developed by Takara (and it's even where the name SMART comes from): [https://www.idtdna.com/pages/education/decoded/article/use-of-template-switching-oligos-%28ts-oligos-tsos%29-for-efficient-cdna-library-construction]

When I asked ChatGPT "are template switching bases that end with ggg part of adapter or not", it said:
Template switching bases that end with "GGG" are typically part of the template-switching oligonucleotide (TSO), not the adapter.
In template switching, an enzyme like reverse transcriptase adds several nucleotides to the 3' end of the cDNA, typically consisting of cytosines (CCC). The TSO, which usually contains a sequence starting with "GGG," hybridizes to these cytosines, allowing the enzyme to continue synthesizing the cDNA. This "GGG" sequence is a crucial component of the TSO and not part of the adapter sequence.
The adapter sequence, on the other hand, is usually added during a later stage of library preparation and is used for subsequent PCR amplification and sequencing. The adapter does not typically include the "GGG" sequence used for template switching.
I wasn't sure what the difference between nontemplated bases and the G bases was, and I wasn't sure if the G bases are part of the TSO or not. But ChatGPT answered that the G bases are part of the TSO but the term "nontemplated bases" refers to the C bases that are added by reverse transcriptase and not to the G bases in the TSO:
- Non-Templated Bases:
- During reverse transcription, the reverse transcriptase enzyme sometimes adds a few non-templated cytosine bases ("C") at the 3' end of the newly synthesized cDNA. This addition is a key part of the template-switching process.
- Template-Switching Oligonucleotide (TSO):
- The TSO is a short synthetic sequence that contains a few guanine bases ("G") at its 3' end. The "G" bases in the TSO are designed to pair with the non-templated "C" bases added to the cDNA.
- Once the non-templated "C" bases are added to the cDNA, the TSO anneals to these bases, allowing the reverse transcriptase to continue copying the TSO sequence into the cDNA.
- Sequence Incorporation:
- The non-templated "C" bases and the complementary "G" bases from the TSO are incorporated into the final cDNA product. As a result, the beginning of your cDNA (and subsequently your sequencing reads) will contain the TSO sequence, often preceded by these non-templated bases.
When I asked ChatGPT how I can trim TSO sequences from the start of
reads that consist of up to 5 bases and that usually end in a G base, it
told me to use fastp --trim_front1=5 (which is exactly what
I did to get rid of the two extra G bases at the start of my MEGAHIT
contig, even though I also trimmed 5 bases from the back end of reverse
reads).
Added in February 2025: USMortality tweeted: "You
know why McKernan's adapter theory is probably also false?! Because why
would this only occur at the ends? If it were true that there are still
technical sequences at the ends of the reads, this should apply to all
reads, implying that we should observe similar mismatches everywhere
else, not only at the ends."
[https://x.com/USMortality/status/1887907383126503722]
But I think it does also occur elsewhere. If you do local alignment and
you keep only reads with soft-clipped bases at the start, the segment of
soft-clipped bases often ends with one or more G bases like here (where
the first column shows the starting position, and in the second column
for example 2S149M means that there are 2 bases at the
start that were clipped by the local alignment):
$ fastq-dump --gzip -X 10000 --split-3 SRR10971381 Read 10000 spots for SRR10971381 Written 10000 spots for SRR10971381 $ minimap2 -a --sam-hit-only sars2.fa SRR10971381_1.fastq.gz >temp.sam 2>/dev/null $ awk -F\\t '$6~/^[0-9]+S/' temp.sam|cut -f4,6,10|column -t|cut -c-80 27223 2S149M GGCAGGTCACTATAGCAGAGATATTACTAATTATTATGAGGACTTTTAAAGTTTCCATTTG # clipped GG 4504 2S130M2S CGGGGTGTGGTTGATTATGGTGCTAGATTTTACTTTTACACCAGTAAAACAACTGTAGCGT # clipped CG 27246 3S128M4S GGGACTAATTATTATGAGGACTTTTAAAGTTTCCATTTGGAATCTTGATTACATCATAAAC # clipped GGG 29254 3S134M GGGGGGAACGTGGTTGACCTACACAGGTGCCATCAAATTGGATGACAAAGATCCAAATTTC # clipped GGG 14080 47S5M1D99M AGCTGGGATTGTTGGTTGTATTTAAATAGTATAAGAACTAATAAATGGGTAATGGTATGAT 17748 3S131M5S CGGTGCTTGGAGAAAAGCTGTCTTTATTTCACCTTATAATTCACAGAATGCTGTAGCCTCA # clipped CGG
In August 2024 USMortality received this response to a FOIA request (the ellipses are part of the original post): [https://www.usmortality.com/p/an-official-cdc-foia-response-confirms]
CDC has responded to my FOIA request...

..., in which I have asked for records related to these four points:
- Records on single virion sequencing of SARS-CoV-2 that ensured the virion was physically isolated from any other genetic material before sequencing.
- Records of a single sequencing (long-)read from the first position [..] to the last position [..] of the genome. [..]
- Records of a perfectly assembled contig [..] by a metagenomic assembler like Megahit (as used by Wu et al, 2020) [..].
- Records of RACE (Rapid Amplification of cDNA Ends) processes carried out on a patient sample or viral culture [..]
In response to the first point, single-virion sequencing is difficult to perform for RNA viruses according to a paper from 2020 titled "Single-virus genomics and beyond": [https://rua.ua.es/dspace/bitstream/10045/110342/5/Martinez-Martinez_etal_2020_NatRevMicrobiol_accepted.pdf, via https://x.com/Agus_Z_X/status/1804624869553770934]
However, SVG has its own biases and technical challenges. For example, the detection of viruses with very small capsid size and/or ssDNA and RNA genomes is virtually impossible with current flow cytometers, which are mostly designed for targeting cells instead of nanoparticles. The low fluorescence derived from the staining of ssDNA and RNA viral genomes with commercially available dyes and their low side and forward scatter signals are below detection limit or overlap with background signal and electronic noise. The development of flow cytometry instruments that can detect very small, low-fluorescence particles would help pushing the boundaries of SVG to capture hidden viral diversity. Other strategies for virus detection and sorting based on microfluidic nano-devices and lab-on a chip with optics integration are becoming very attractive alternatives to flow cytometry. Despite the well-established protocols for detecting and targeting dsDNA viruses using fluorescent nucleic acid-binding stains, to the best of our knowledge, current commercial dyes with high affinity for ssDNA and RNA do not fully discriminate against dsDNA, further complicating the distinction of ssDNA and RNA containing particles. Furthermore, although Phi29 DNA polymerase and variations of it can amplify DNA, we are not aware of a commercially available enzyme with the required sensitivity for whole genome amplification (WGA) of a single copy of a RNA virus genome.
In response to the second point, even though there wouldn't be a sequencing run where the entire genome had been sequenced in a single read, there are direct RNA nanopore sequencing runs which contain reads that are over 10,000 bases long, like for example SRR11300652 which I described in an earlier section. And even in the case that someone would've managed to sequence nearly the entire genome in a single read, the CDC wouldn't necessarily have any records relating to the study if the study wasn't performed by the CDC.
In response to the third point, in previous sections of this file I have explained why MEGAHIT contigs don't include the full 33-base poly(A) tail and why they sometimes include one or two extra G bases of the 5' end.
The CDC responded to the 4th point by saying that they haven't used 5' RACE to sequence SARS-CoV-2. However there are many other institutions that have used 5' RACE to sequence SARS-CoV-2.
USMortality claimed that the FOIA response "confirms that the validation of the SARS-CoV-2 genome has not been completed to scientific standards". [https://x.com/USMortality/status/1825961166667133203] However the validation of the genome is not a scientific concept but it's a concept that was invented by USMortality. For example one of his criteria for establishing the validity of the genome is valid is that he finds ten or more sequencing reads which match the 5' end of the genome perfectly so that no bases have to be trimmed from the ends of the reads. But I don't know any scientific source which has proposed similar criteria for validating the genome of a virus. And single-virion sequencing is a rare technique which currently seems to be difficult or impossible to perform for RNA viruses, so few actual virologists would probably agree that it's necessary to perform single-virion sequencing of SARS-CoV-2 in order to validate the genome to "scientific standards" (which are in fact not scientific standards but standards that were invented by USMortality).
Added in January 2025: USMortality also sent a similar request to the Robert Koch Institute which is the German equivalent of the CDC, but he received the following response which explained that his questions were stupid: [https://x.com/USMortality/status/1876650223440740355/photo/1]

In this Japanese paper the authors developed a technique which enabled them to sequence the complete 5' and 3' ends of SARS-CoV-2 without having to use RACE: [https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2023.1137086/full]
However, the NGS techniques involve a significant burden for sample preparation. Since to generate complete viral genome coverage, genomic nucleic acid enrichment is required by reverse transcription PCR using virus-specific primers or by viral particle concentration. Furthermore, conventional NGS techniques cannot determine the 5' and 3' terminal sequences of the RNA viral genome. Therefore, the terminal sequences are determined one by one using rapid amplification of cDNA ends (RACE). However, since some RNA viruses have segmented genomes, the burden of the determination using RACE is proportional to the number of segments. To date, there is only one study attempting whole genome sequencing of multiple RNA viruses without using above mentioned methods, but the generated sequences' accuracy compared to the reference sequences was up to 97% and did not reach 100% due to the low read depth. Hence, we established novel methods, named PCR-NGS and RCA-NGS, that were optimized for an NGS machine, MinION. These methods do not require nucleic acid amplification with virus-specific PCR primers, physical viral particle enrichment, and RACE. These methods enable whole RNA viral genome sequencing by combining the following techniques: (1) removal of unwanted DNA and RNA other than the RNA viral genome by nuclease treatment; (2) the terminal of viral genome sequence determination by barcoded linkers ligation; (3) amplification of the viral genomic cDNA using ligated linker sequences-specific PCR or an isothermal DNA amplification technique, such as rolling circle amplification (RCA). The established method was evaluated using isolated RNA viruses with single-stranded, double-stranded, positive-stranded, negative-stranded, non-segmented or multi-segmented genomes. As a result, all the viral genome sequences could be determined with 100% accuracy, and these mean read depths were greater than 2,500×, at least using either of the methods.
The methodology used in the paper was demonstrated in this figure:
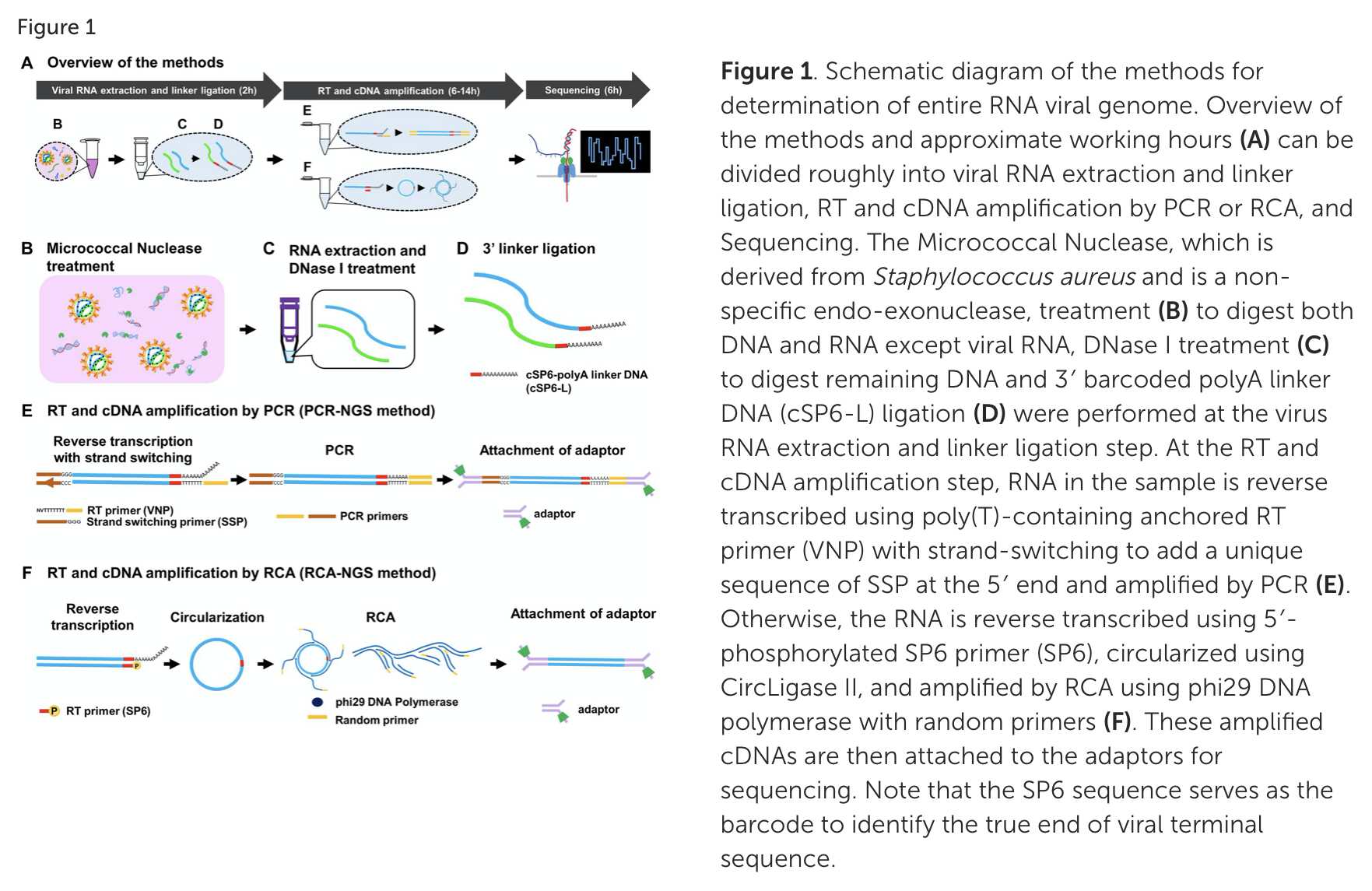
When I tried aligning the reads for SARS-CoV-2 from the paper against
Wuhan-Hu-1, the reads with starting position 1 all included a sequence
of about 100 bases before the actual read. The sequence generally seem
to end with 2 to 5 G bases similar to Wu et al.'s reads,
which are part of the template switching oligo which anneals to the
complementary untemplated C bases that are added by reverse
transcriptase:
seqdif()(seqkit fx2tab|gawk -F\\t '{name[NR]=$1;split($2,z,"");for(i in z)seq[NR][i]=z[i];if(length($1)>max1)max1=length($1);if(length($2)>max2)max2=length($2)}END{for(i=1;i<=max2;i+=width){printf("%"(max1+1)"s","");for(pos=i;pos<i+width&&pos<=max2;pos+=10)printf(pos-i>width-10?"%s":"%-10s",pos);print"";for(j in seq){out=sprintf("%"max1"s ",name[j]);for(k=i;k<=max2&&k<i+width;k++)out=out (seq[j][k]==seq[1][k]?seq[j][k]:"\033[31m"seq[j][k]"\033[0m");print out}}}' "width=${1-60}")
curl "https://www.ebi.ac.uk/ena/portal/api/filereport?accession=DRR428582&result=read_run&fields=fastq_ftp"|sed 1d|cut -f2|tr \; \\n|sed s,^,ftp://,|xargs wget
curl 'https://eutils.ncbi.nlm.nih.gov/entrez/eutils/efetch.fcgi?db=nuccore&rettype=fasta&id=MN908947'>sars2.fa
bowtie2-build sars2.fa{,}
bowtie2 --no-unal -p4 -x sars2.fa -U DRR428582_1.fastq.gz>DRR428582.sam
awk '$4==1' DRR428582.sam|head|cut -f1,10|seqkit tab2fx|cat <(seqkit replace -p'.* ([^ ]*),.*' -r\$1 sars2.fa) -|seqkit subseq -r 1:1000|mafft --quiet --thread 4 -|seqdif 80

The reads also end with a sequence that is about 100 bases long, where based on the figure in the screenshot above I guess the first part consists of a PCR primer and the second part consist of an adapter. The poly(A) tail seems to be about 10-20 bases long in most reads:
samtools view DRR428582.sam|awk '$4>=28000'|head|cut -f1,10|seqkit tab2fx|cat <(seqkit replace -p'.* ([^ ]*),.*' -r\$1 sars2.fa) -|seqkit subseq -r -1000:-1|mafft --quiet --thread 4 -|seqkit subseq -r -160:-1|seqdif 80

In the approximately 100-base sequences at the start and end of reads, the outer half seems to be more variable but the inner half is more constant, but it might just be if base calling errors are more common at the ends of reads, because there is low average quality in approximately the first 50 and last 50 bases of the reads:

Based on this figure from the Japanese paper, the inner half of the 5' end seems to correspond to the brown segment (strand switching primer) and the outer half seems to correspond to the light purple segment (adapter):
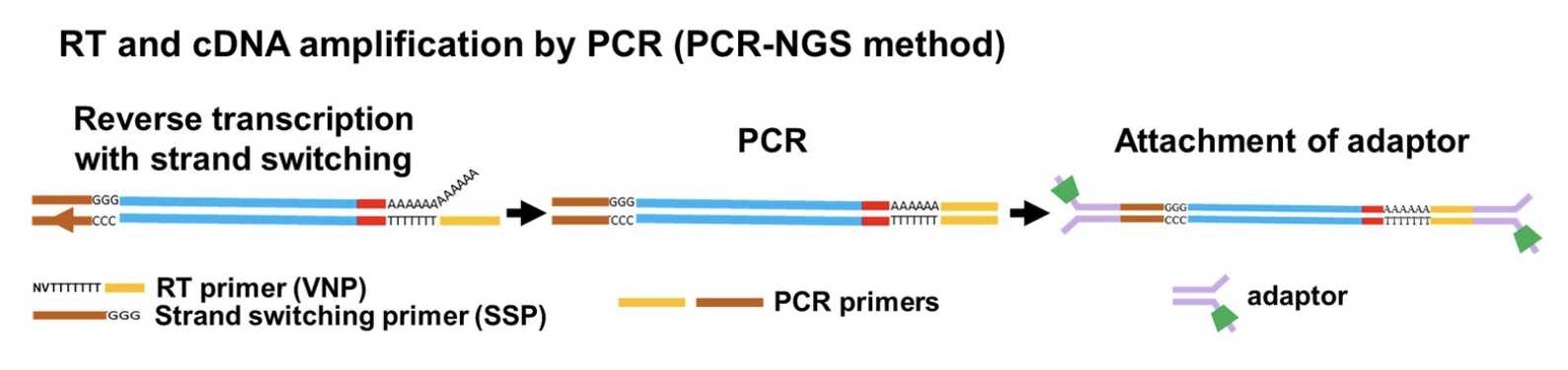
The longest read that aligned against Wuhan-Hu-1 was 8,454 bases long, but the first 111 and last 52 bases were clipped by BLAST. It has only about 90% identity to Wuhan-Hu-1, but a 10% error rate is typical for the ONT MinION sequencer which was used in the study:
$ makeblastdb -dbtype nucl -in sars2.fa [...] $ fmt='pident qcovs qlen qstart qend sstart send';samtools fasta DRR428582.sam|seqkit sort -lr|seqkit head -n1|blastn -db sars2.fa -outfmt "6 $fmt"|(echo $fmt;cat)|column -t pident qcovs qlen qstart qend sstart send 89.885 98 8454 112 8402 21321 29890